Replication, repair and combination
There are "traffic lights" controling DNA replication. (Tuesday)
Whats the emergency stops the DNA replication (Thursday)
Control of Replication in Bacteria
Focusing on how bacteria initiates and terminates
Origins of replication are AT-rich and are bound by initiator proteins.
The origin area is the replicator, which is interacted with initiator protein.
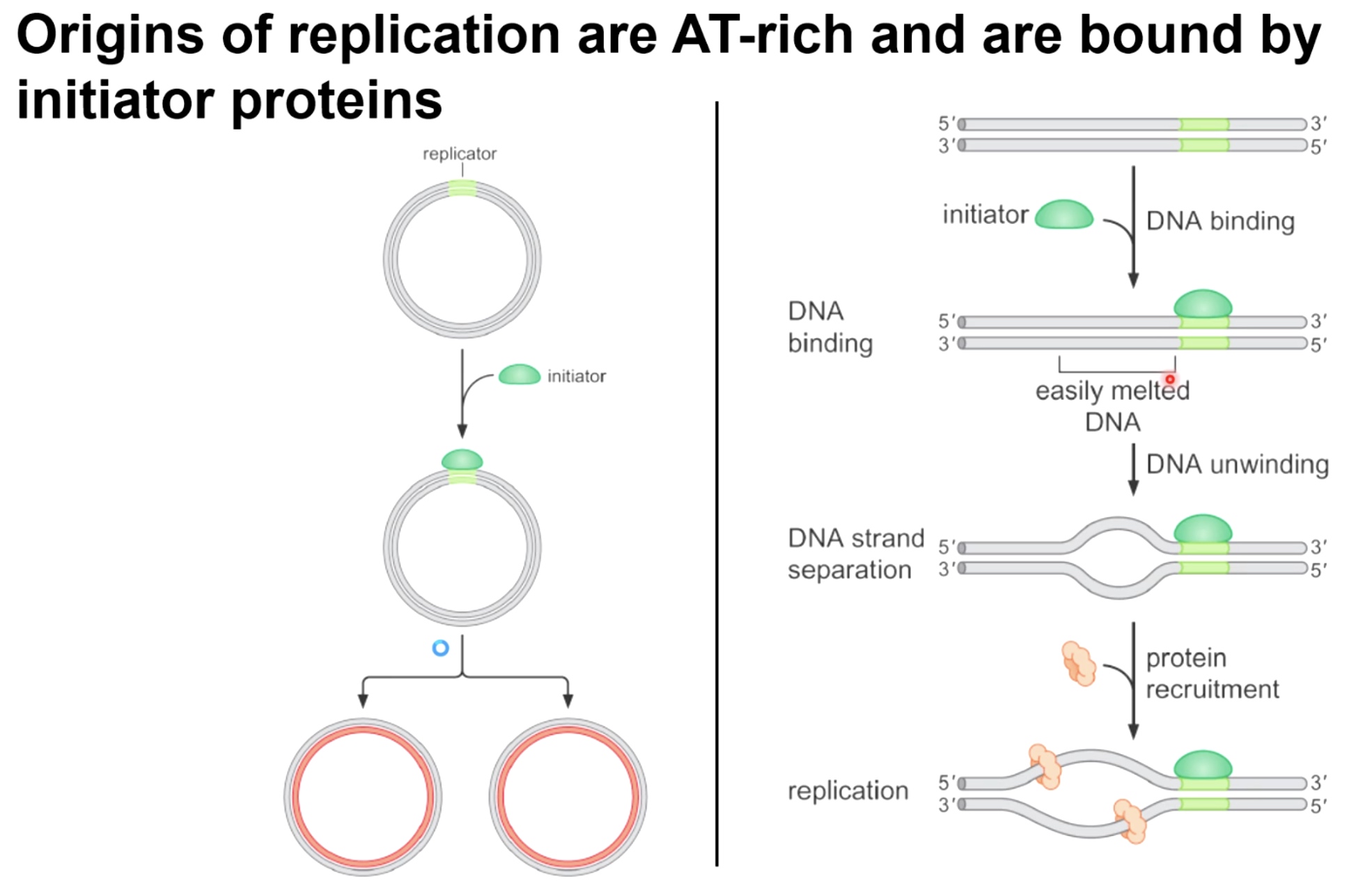
Initiator is going to bind specific DNA regions which are AT-rich and are easily melted, which is going to allow for a concerted DNA unwinding effort, which creates single stranded DNA, where the DNA helicase can recruited to and can bind and circling that isngle-strand DNA.
Strategies for Initiation of Replication
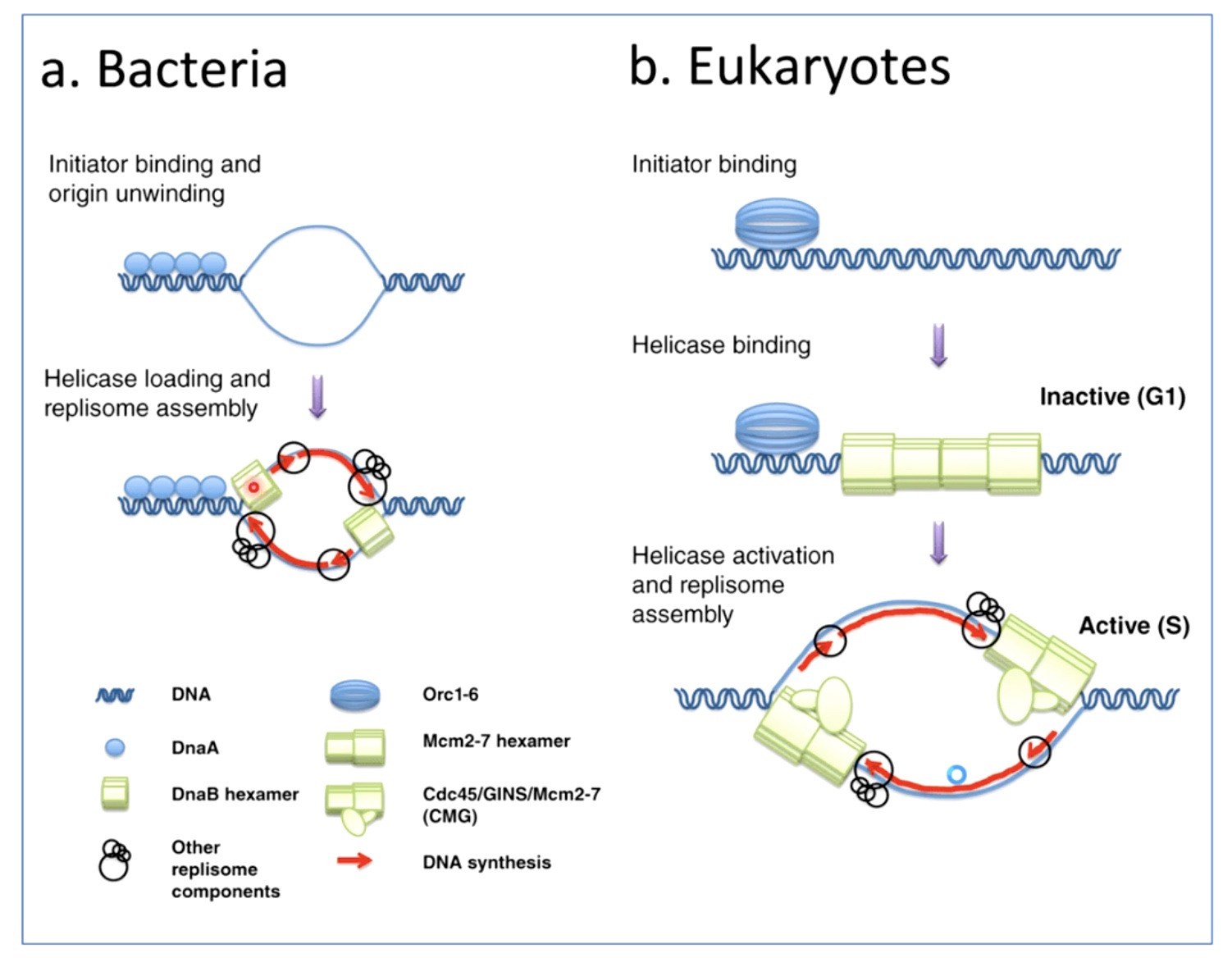
Bacteria and Eukaryotes do the initiation of replication very differently.
Bacteria
When the initistor bind, the origin unwinds almost immediately, helicase can load.
Eukaryotes
The initiator binds, and it allows the helicase to bind as well. But the helicase has to be activated by certain cellular signals and tell the helicase is activated. The helicase remains inactive until it's time for DNA replication to happen during phase down.
Initiation of Replciation in E.coli
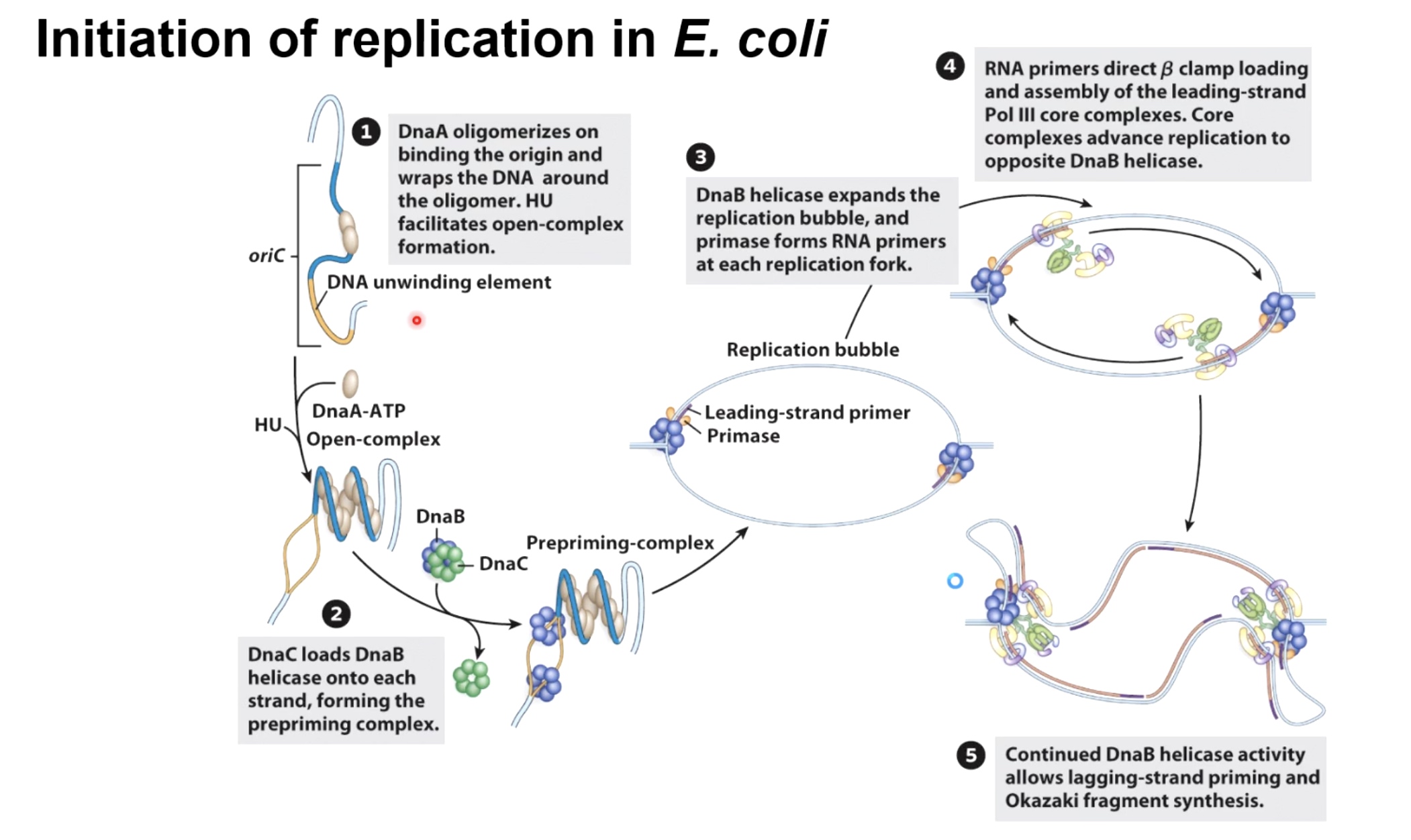
The initiate protein in bacteria is called DnaA It goes to wrap the DNA around it.
Then the protein called HU, is going to cause the formation of an open-complex.
The helicase loader complex is called DnaC in bacteria, load the homoheximer DnaB helicase on each side of growing replcation forks. They will initiate unwinding to form the prepriming complex.
Helicase then use hydrolysis of ATP to move away form each other, recruit the primase, which will lay down the leading-strand primers. The replication bubbles will continue to open up.
Initially, with only the leading strand being synthesized. RNA primers direct beta clamp loading and assembly of the leading-strand pol III core complexes. Core complexes advance replication to opposite DnaB helicase.
Continued DnaB helicase activity allows lagging-strand priming and Okazaki fragment synthesis.
Structure of the E.coli Origin
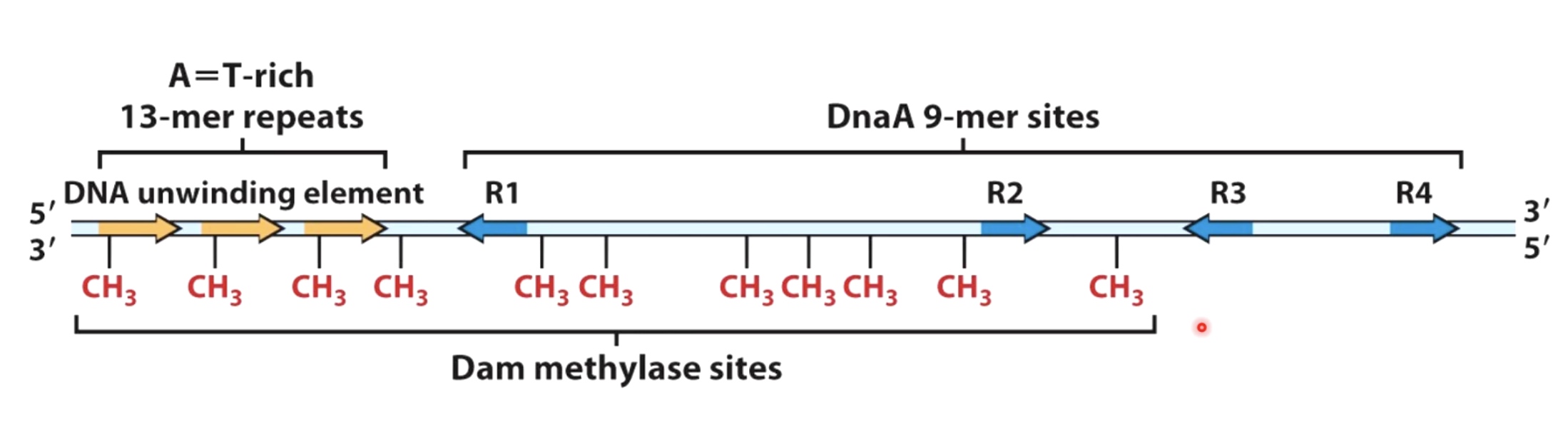
Initiator will bind specifically to the DnaA 9-mer sites.
These sites are substrates for enzyme called dam methylase. It will recoginze GATC repeats and it will methylated on the adenine in those GATC repeats.
Replication Initiation in E.coli depends on DNA methylation Status
Fully methylated DNA
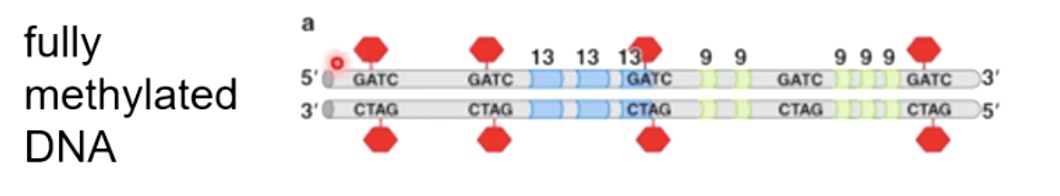
DnaA will only bind to fully methylated DNA. And the binding will allow DNA to open up.
As a new strand being synthesized, it takes a while for them to be methylated by a dam methylase.
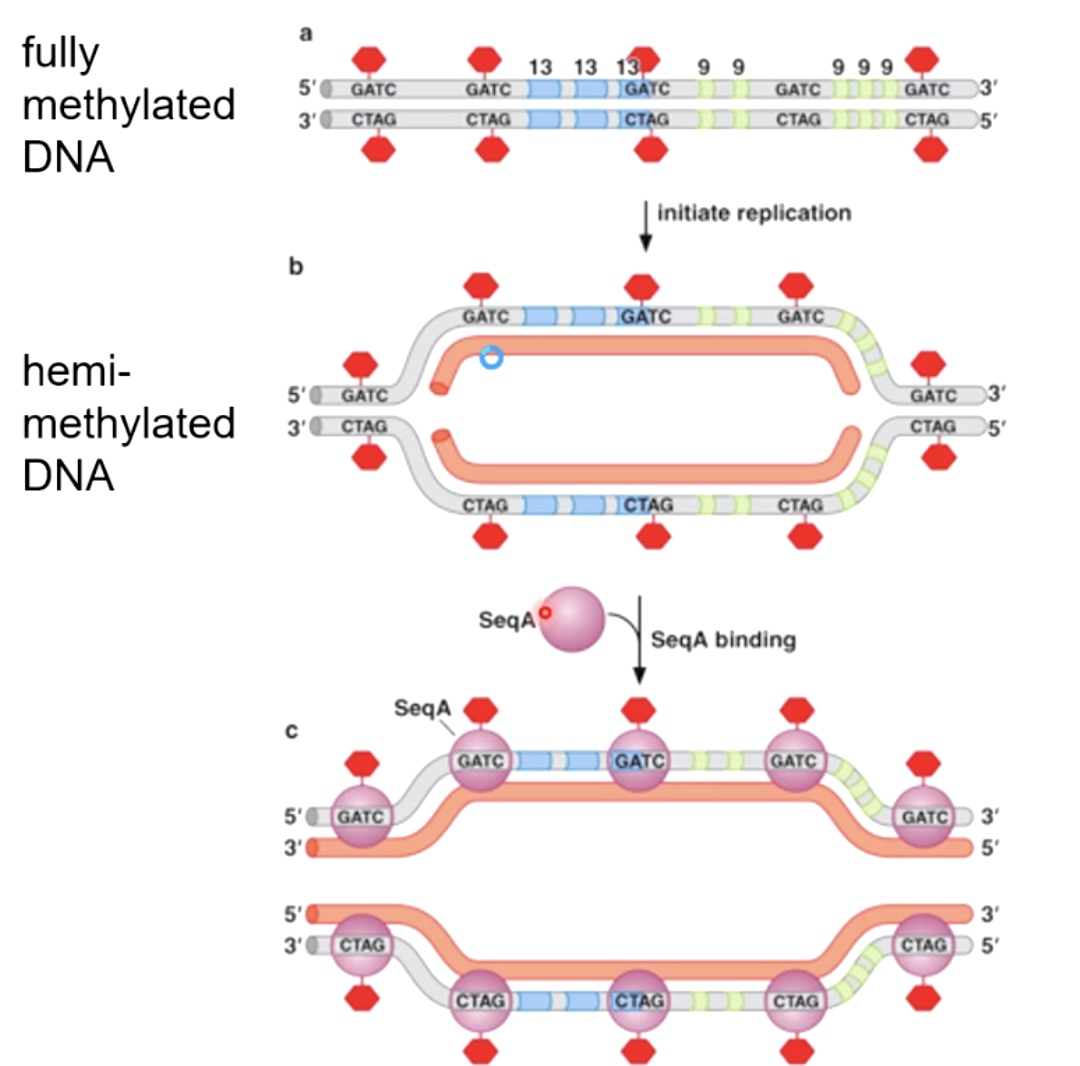
Hemi-methylated DNA is an excellent substrate for binding a protein called SeqA. SeqA binds with high affinity to hemi-methylated DNA.
What SeqA does is it prevents the action of dam methylase from coming in and methylate the other strand. By prevent the action of dam methylase, which in turn prevents the sequence specific binding of DnaA.
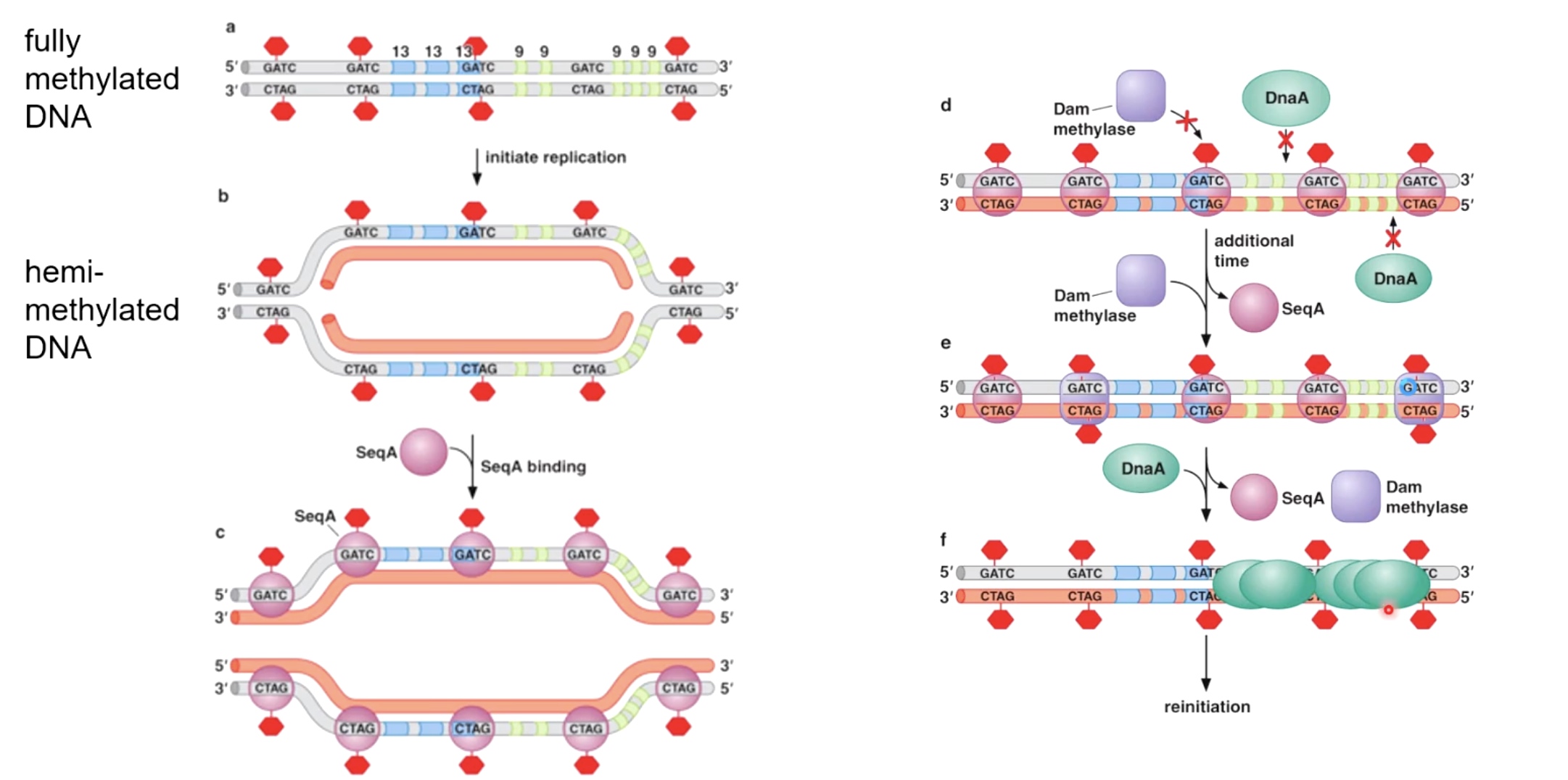
After SeqA disassociated with DNA, the dam methylase begin to sneak in and methylatet the strand. After SeqA leave, the DnaA will come in to reinitiate the DNA replication.
In bacteria, the whole proecess takes between 15 and 20 minutes for Dna to go from hemi-methylated to fully-methylated. The reason is that in bateria, it takes at least 20 minuts to fully copy the circular Dna chromosome. So we do not want to initiate replication before most copying has completed of previous molecule.
Termination of Replication in Bateria
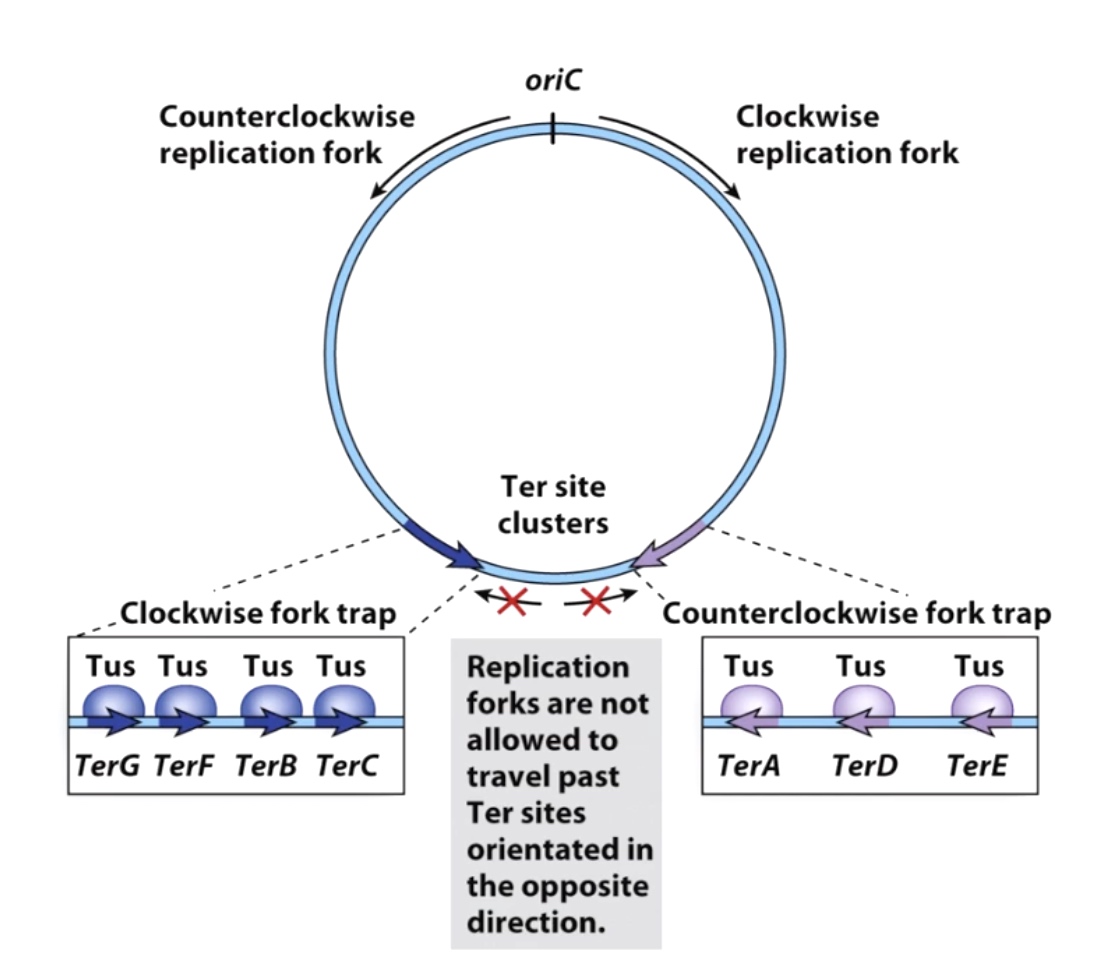
oriC stands for the origiin of the replication. Because replication is bidirectional, so the replication fork will move in opposite directions.
The replicaion fork will going to be trapped by fork trap. The protein called Tus combines specificaly to the TerA, TerD and TerE. The Tuses becomes barrier prevents the freight train of replication. They are polar blocker(one-way barrier), which meas they will block anything that's coming in the counter clockwise direction, but they will allow the clockwise replication forks to go past them.
Sometimes the replciation forks are going to meet, when that happens, there are specialized machinary, including DNA topoisomerases that will untangle the replicated chromosomes and allow the replication machinery to fall off in an oragnized fashion.
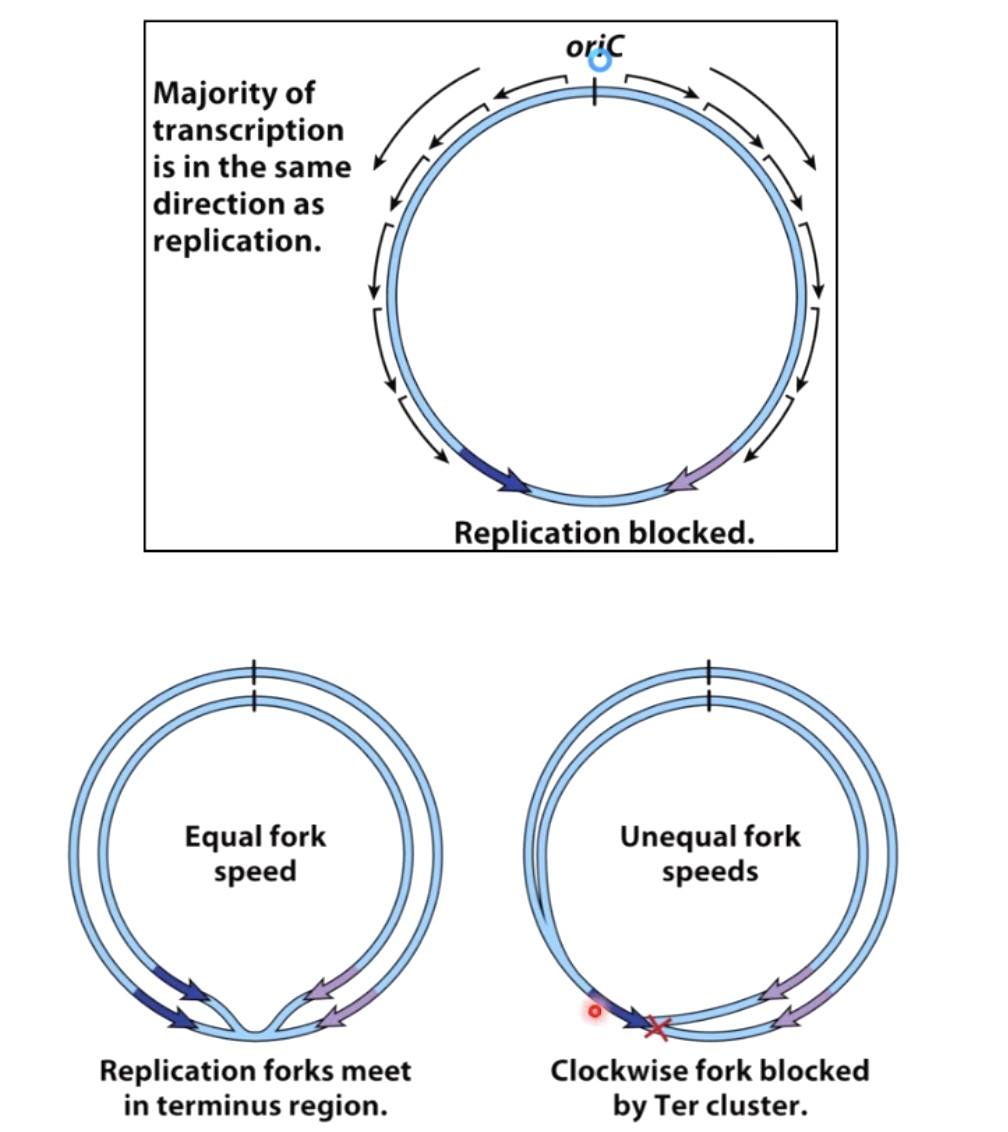
Why should we set the trap?
In bacteria, most of transcription is in the same direction as replication. The worst things happen to a DNA molecule is replication fork moving in one direction and RNA polymerase complex transcribing DNA moving in the opposite direction.
Mismatch Repair
DNA Damage and Inaccurate Replication Can Lead to Mutations
- In bacteria, Polymerase III has a 10-7 error rate.
- Errors also occur when polymerases copy damaged templates
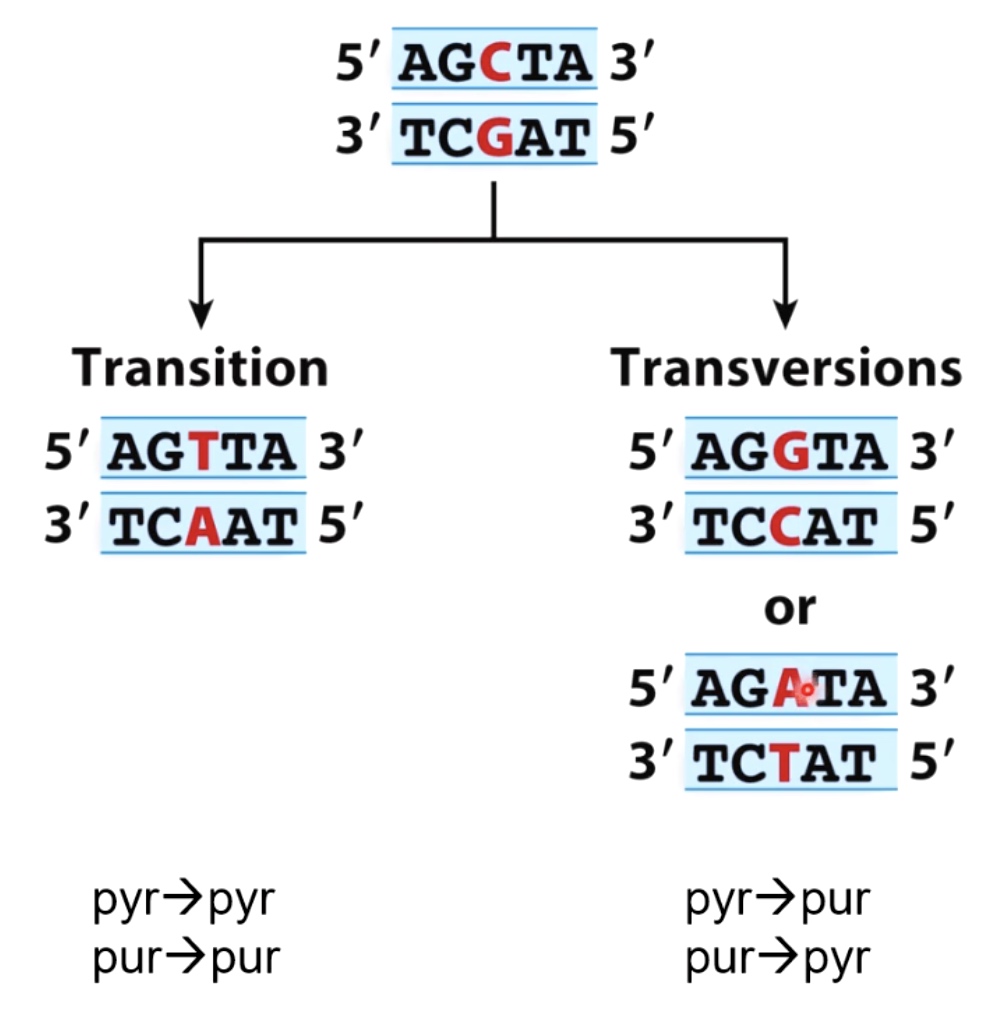
Mutations:
- Transition(pyrimidine -> pyrimidine, purine -> purine)
- Transversions(pyrimidine -> purine, purine -> pyrimidine)
Tautomeric Shifts in Bases can Lead to Replication Errors and Mutations
When DNA is replicating, we can get miss incorporation of incorrect bases, opposite the other strand.
We can see from the picture that th erare enol tautomeric form of guanine is now base pairing with thymine. This GT mismatch commit to a mutation when the cells go on and divide in future replication
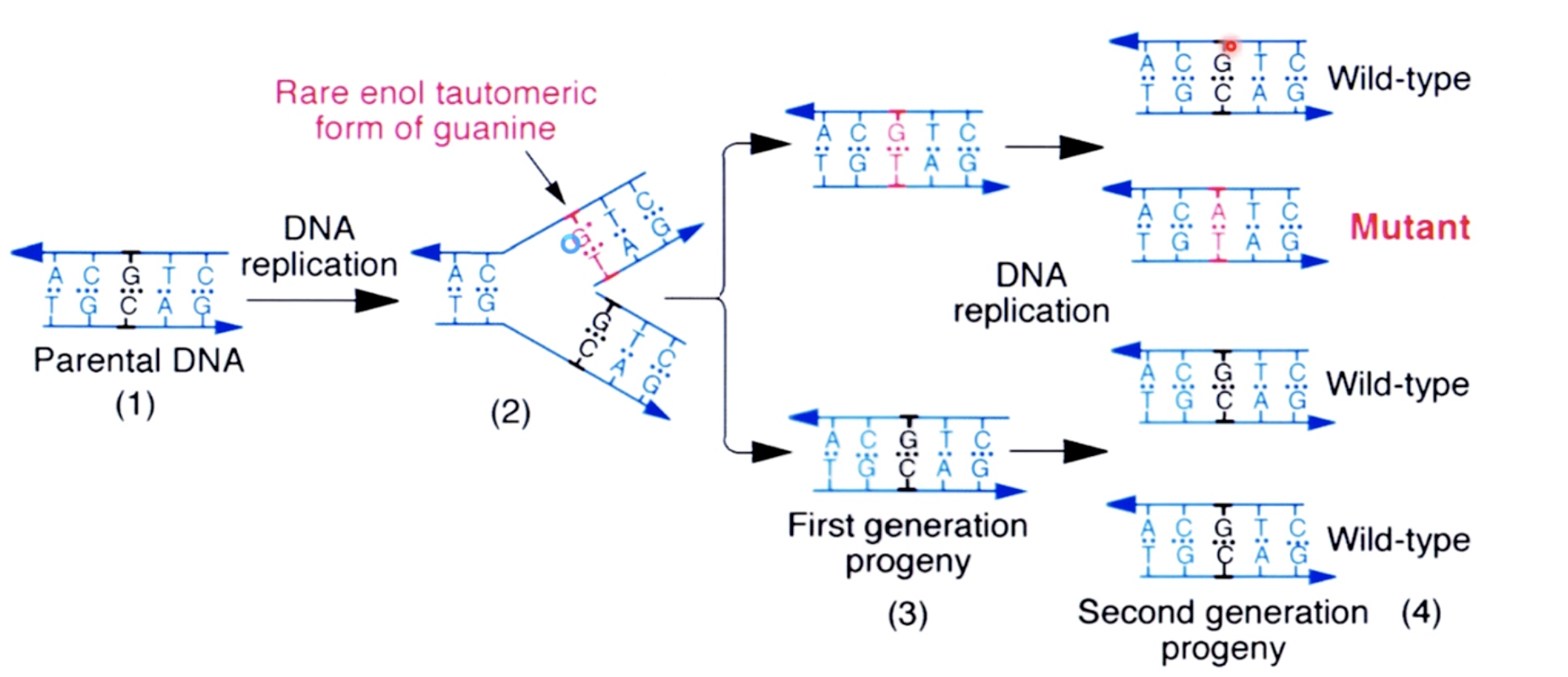
This fullfil the transition from GC to AT.
Mismatch Repair in E.coli
Cells have evolved pathways called mismatch repair which will recognize these mismatches and remove them. Resynthesize the DNA so that the base pairs are correct.
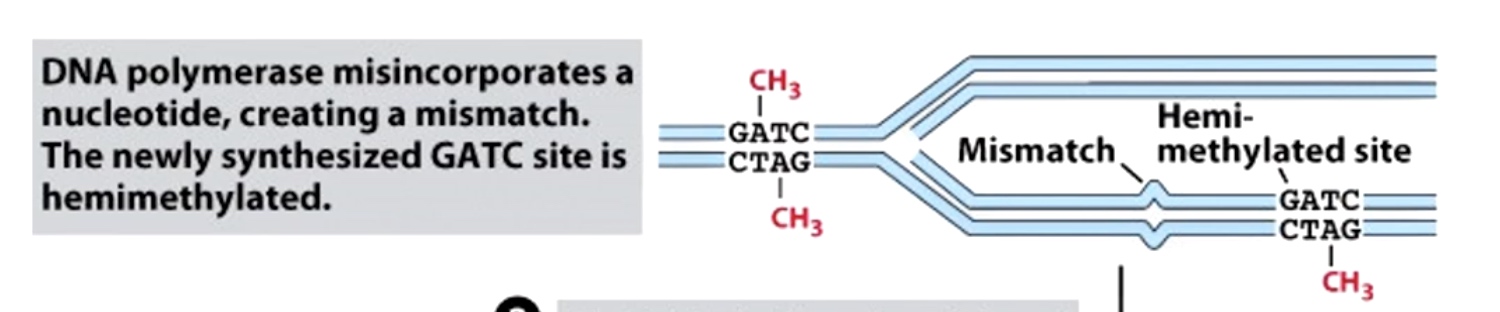
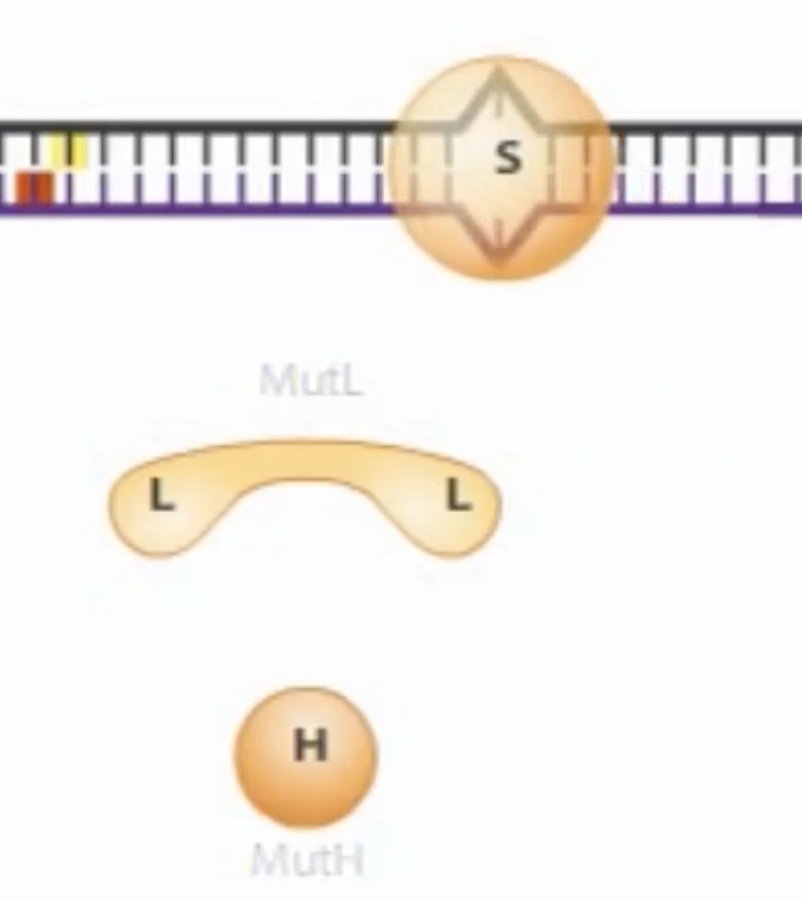
Hemi-methylation DNA is also used during mismatch repair to identify new DNA strands. In mismatch repair, the mismatch is detected in protein called MutS. The MutS binds the misatch and forms a complex with MutL.
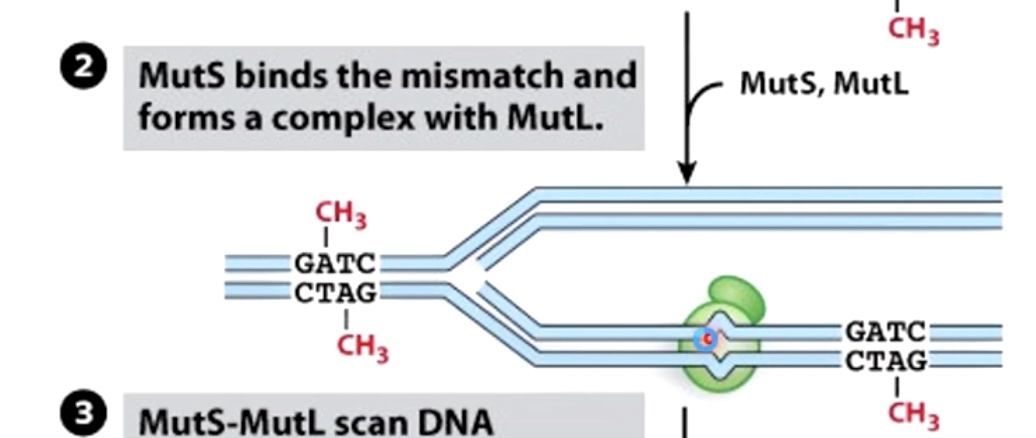
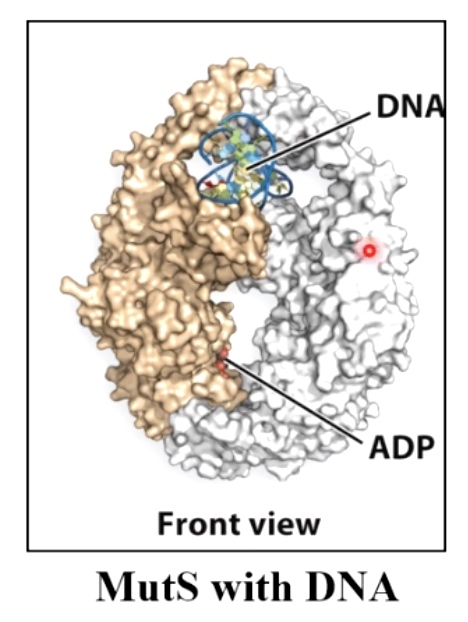
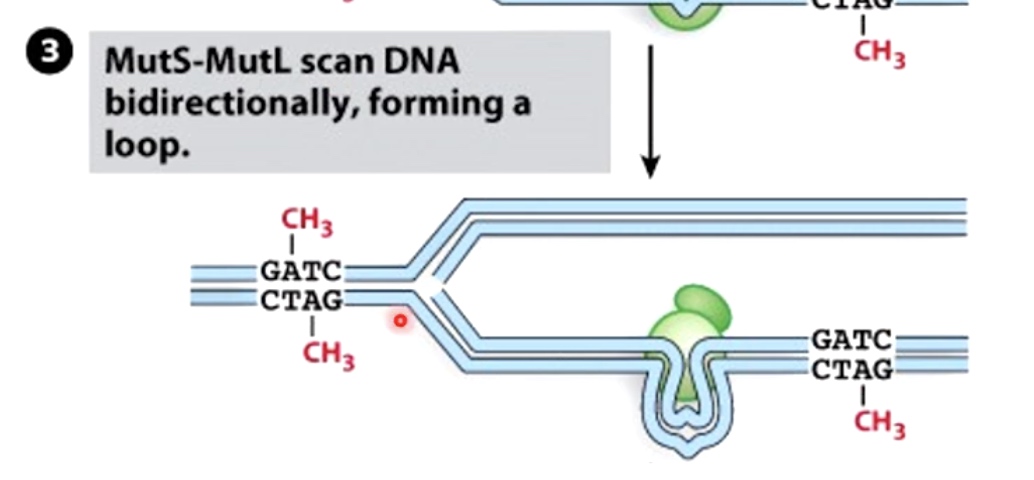
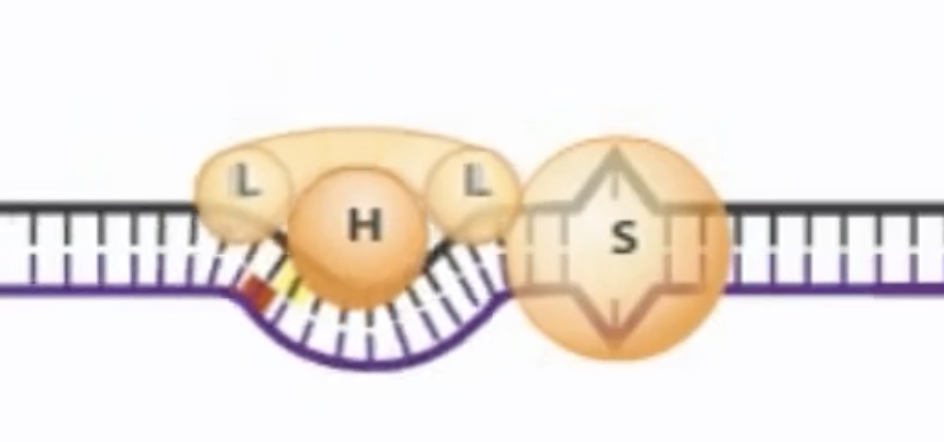
MutS-MutL then scan DNA bidirectionally, forming a loop. It is looking for the GATC hemi-methylated sides.
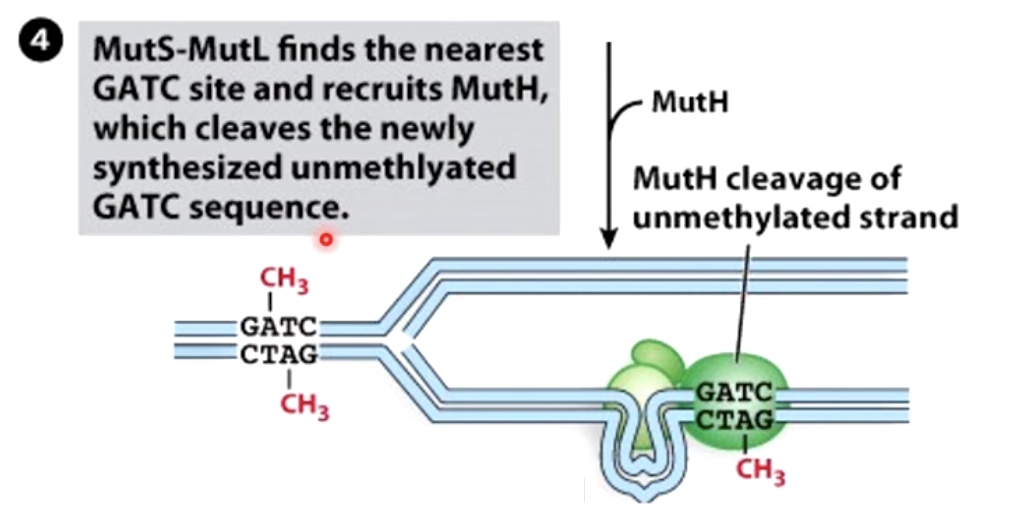
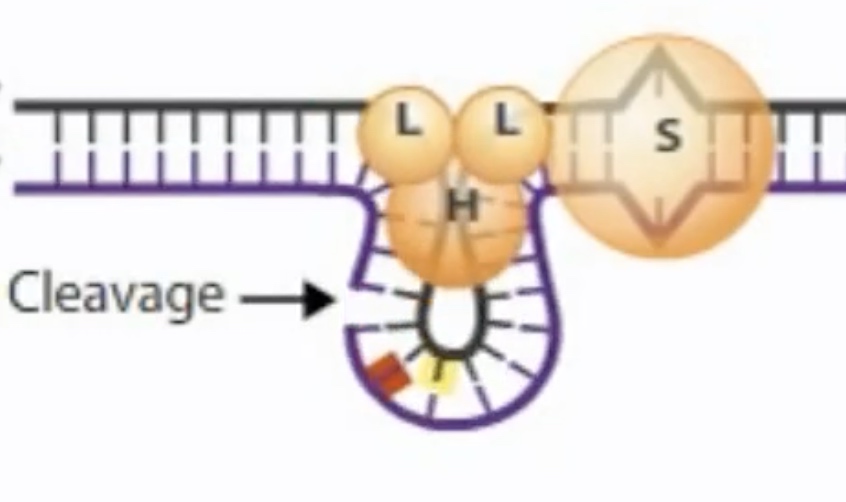
MutS-MutL finds the nearest GATC site and recruits MutH, which cleaves the newly synthesized unmethylated GATC sequence.
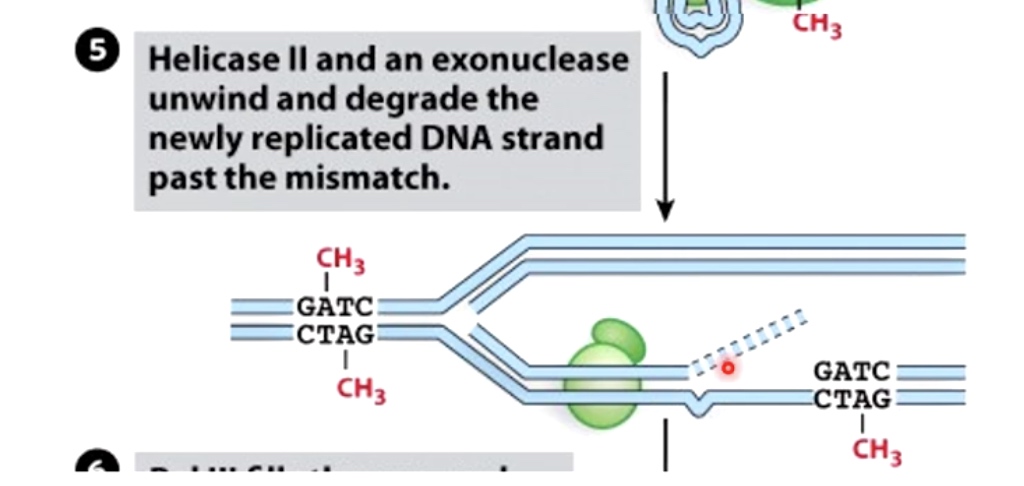
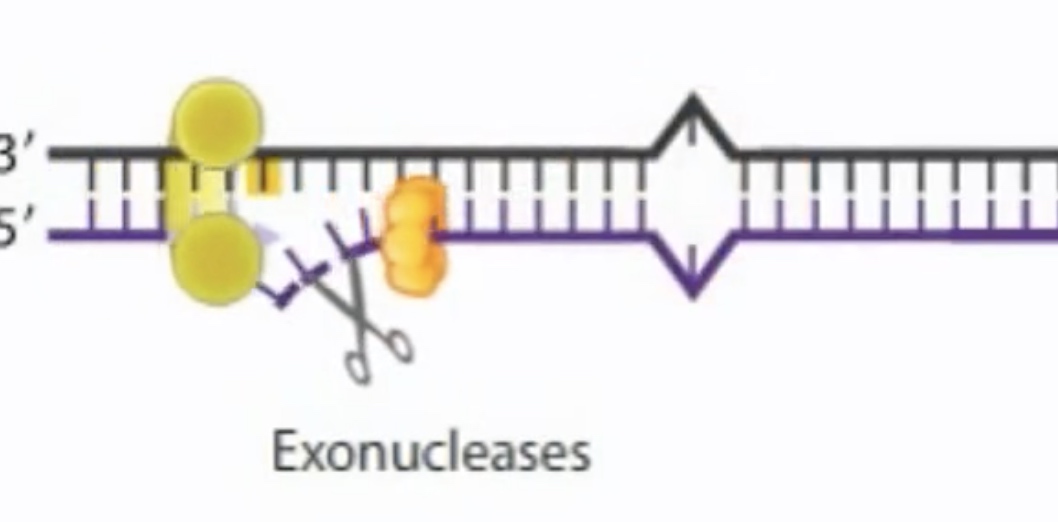
Helicase II and exonuclease unwind and degrade the newly replicated DNA strand past the mismatch.
Endonuclease and Exonuclease
endonucleases cleave the phosphodiester bond in the polynucleotide present internal in the polynucleotide chain, whereas exonucleases cleave the phosphodiester bond from the ends.
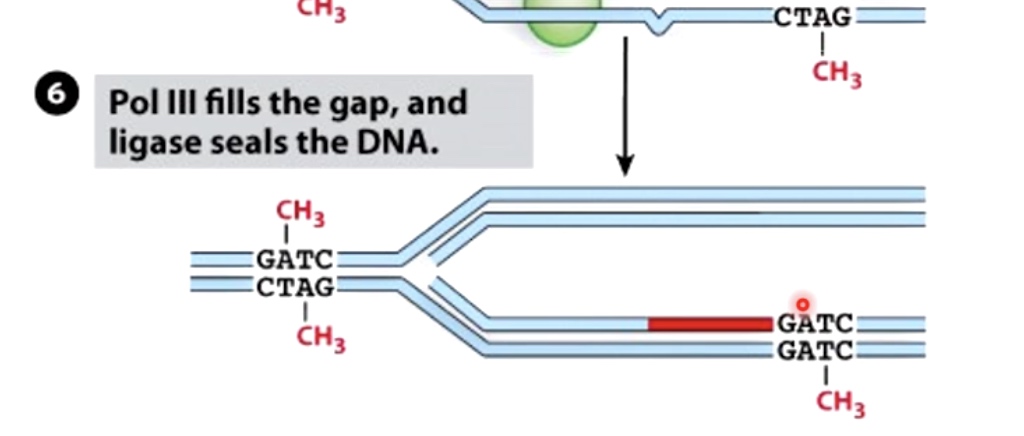
Polymerase III fills the gap, and ligase seals the DNA.
Overview of Mismatch Repair
- Detect mismatch
- Use methylation cues to cleave new DNA strand
- Remove mismatch
- Fill in gap and ligate
Mismatch Repair in Eukaryotes
MSH = MutS homolog
MLH = MutL homolog
Instead of MutH, PMS2 makes the nick.
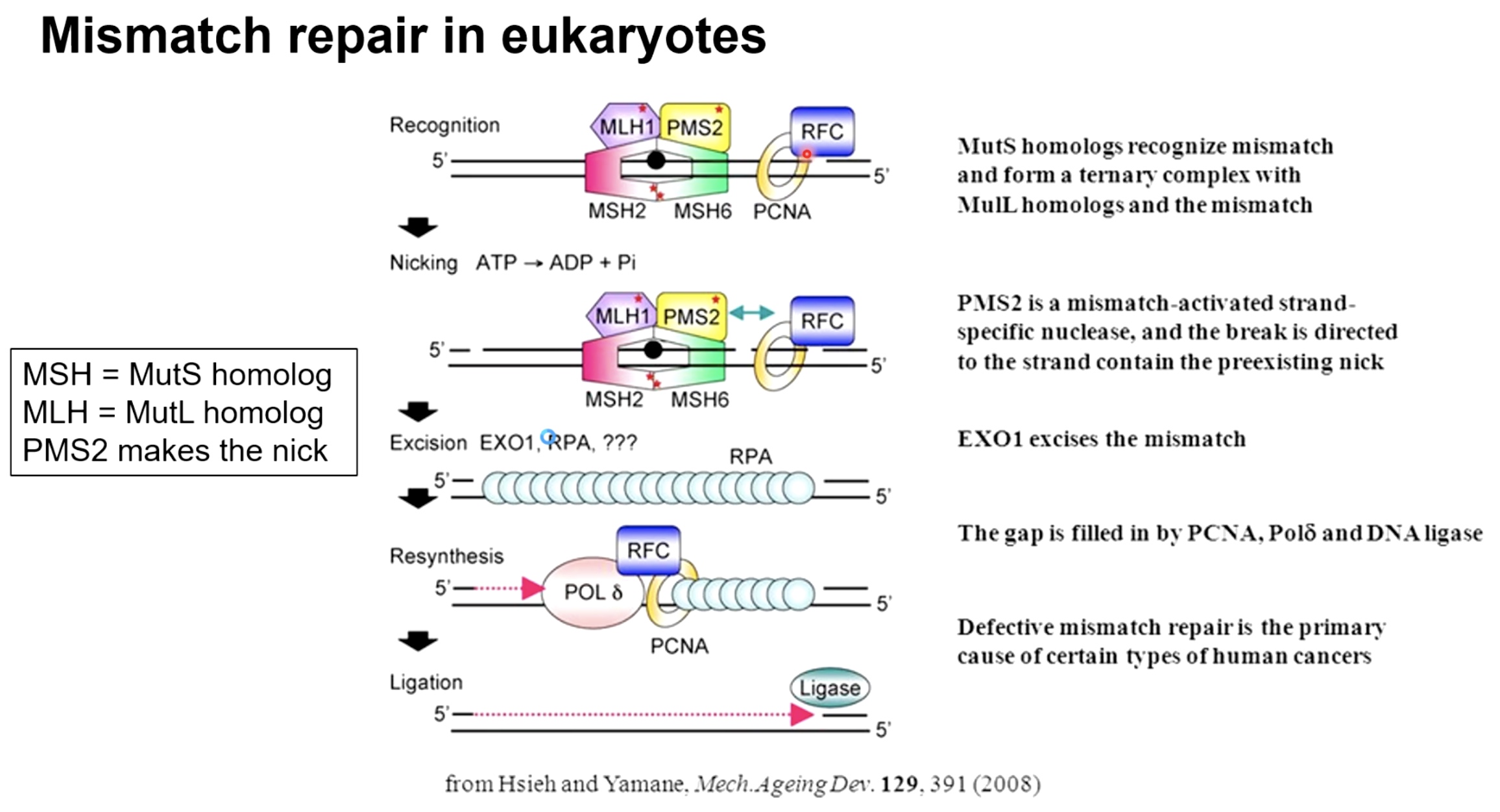
There is on methylation cues in Eukaryotes.
The new different way to detect newly synthesized strand.
- Mismatch orrucing on lagging strand: we have nickes in the DNA prior to when Okizaki fragments are ligated together. And these nicks indicates that this is the strand that is newly syntheszed.
- Leading strand is a little bit more difficult. PCNA sliding clamp is asymmetrically placed on DNA, gives clues as to which of the strands is newly synthesized and which of the strands is the old DNA strand.
The next process contimues the same as in bacteria.
Recogition of msimatch --> nicky --> removal of mismatch strand with exonucleases --> resynthesize
Defective mismatch repair is the primary cause of certaintypes of human cancers.
Mismatch Repair Deficiancy in Humans can Lead to Cancer
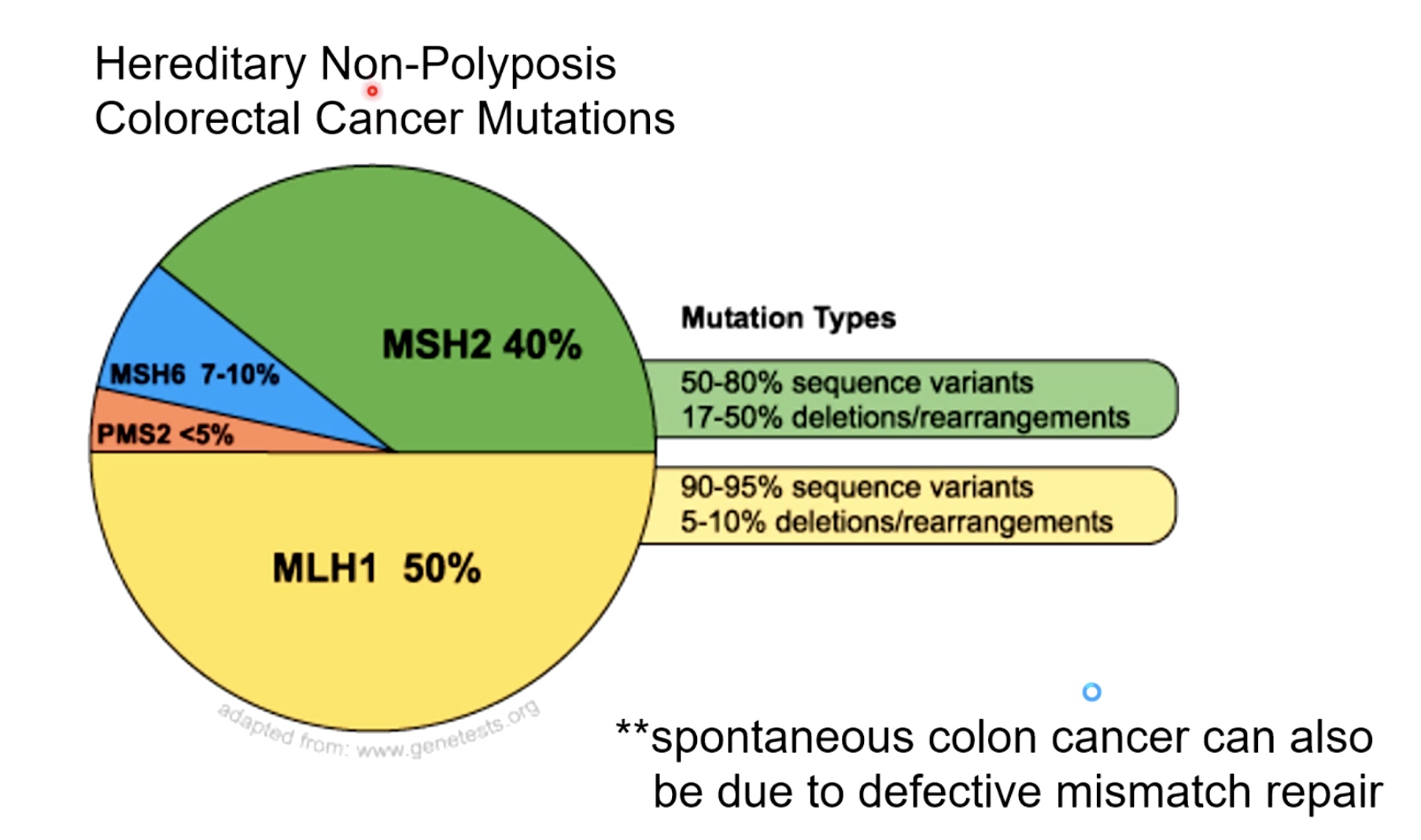
Cancer HNPCC. A large majority of the mutations occur in the mismatch repair protein.
Mismatch Repair Animation
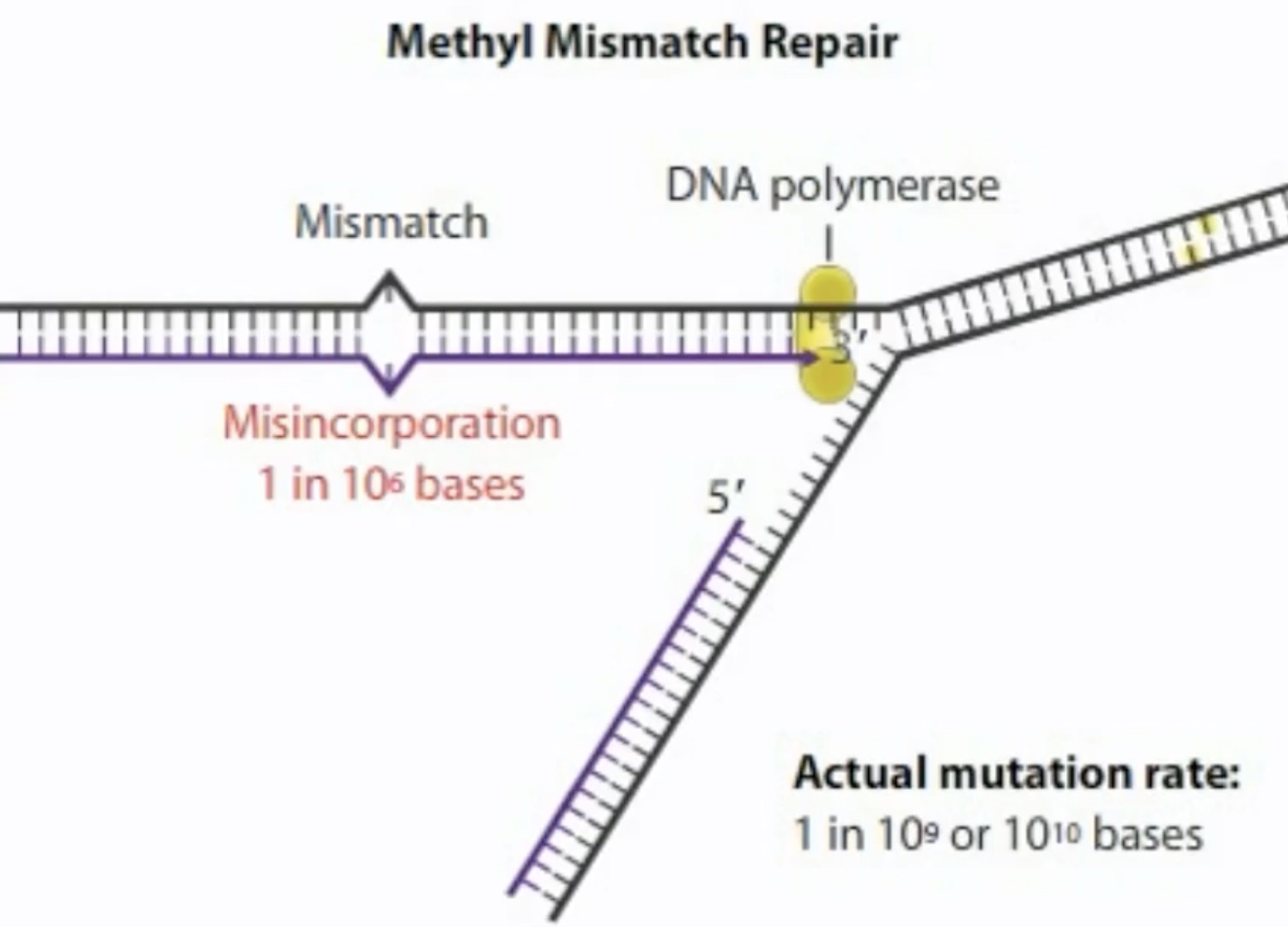
The misincorporation rate us 1 in 1o6 bases. But, the mutation rate in an E.coli cell is only one for every 10^9 or 10^10 bases replicated.
The cell has a number of repair methods, including one called methyl mismatch repair, to remove and replace a mismatched base.
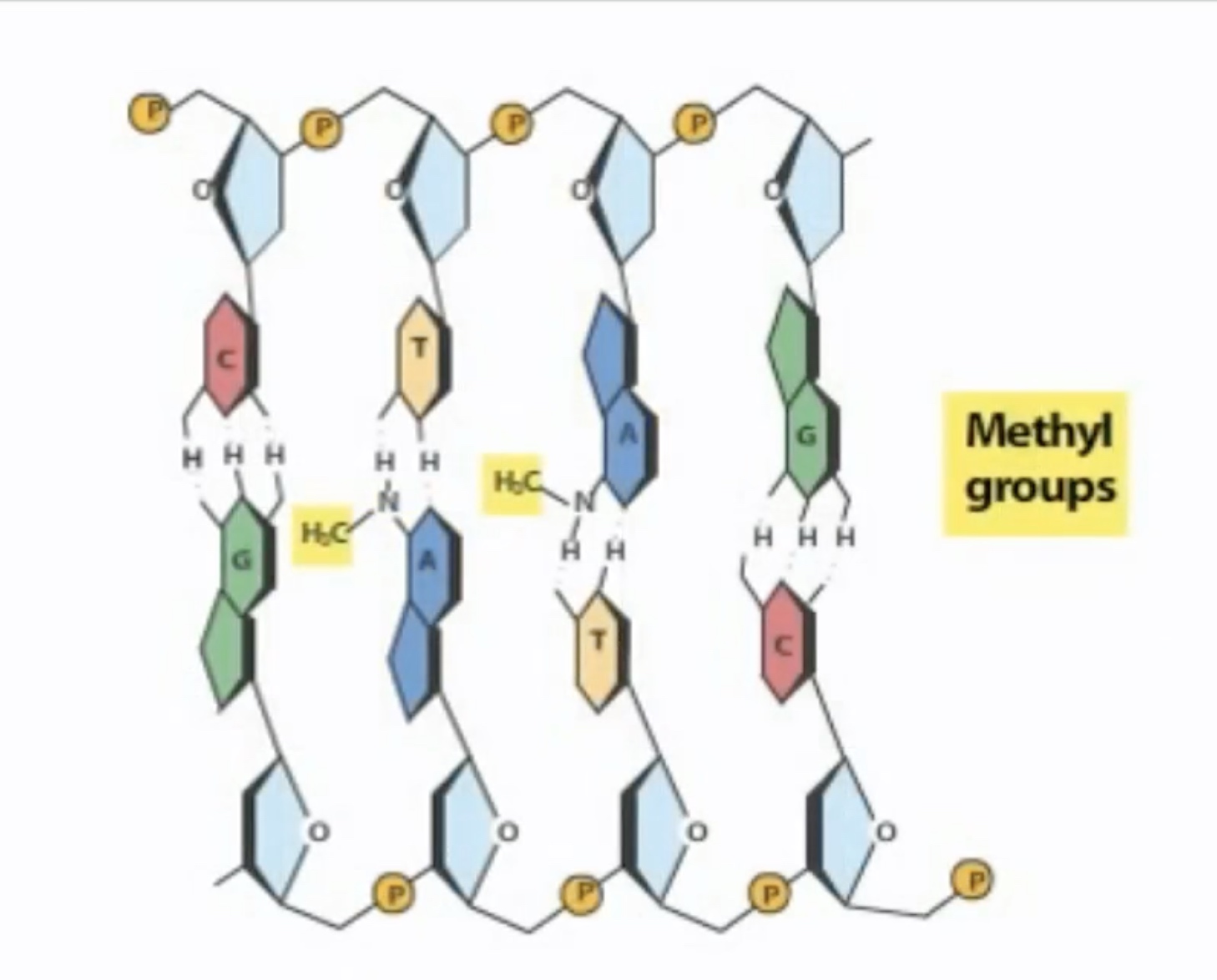
Dam methylase adds a methyl group to the adenine in both strands of the seqiemce.
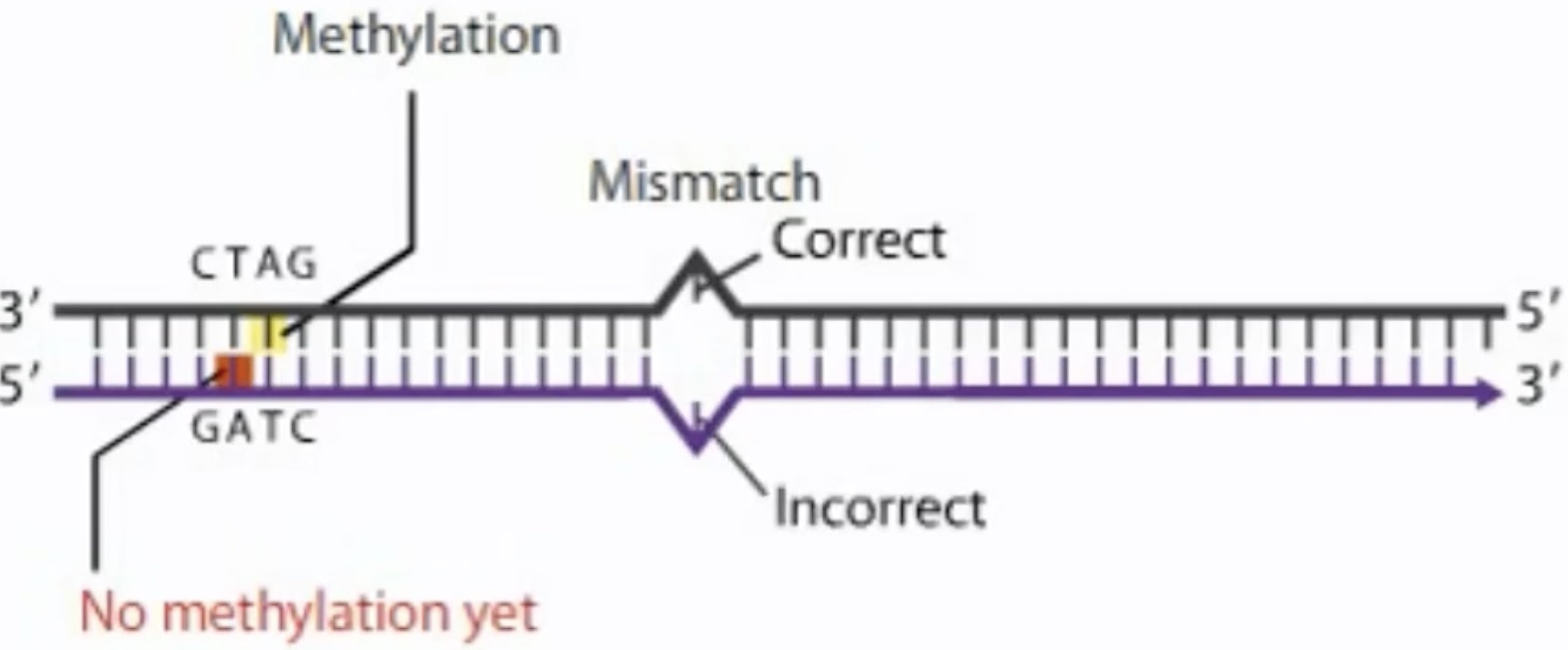
Where the DNA has methylation sequences, the parental strand will be methylated, but the new strand will still be unmethylated. The difference in methylation allows the cell to distinguish the correct sequence on parental strand from the incorrect sequence on the new strand.
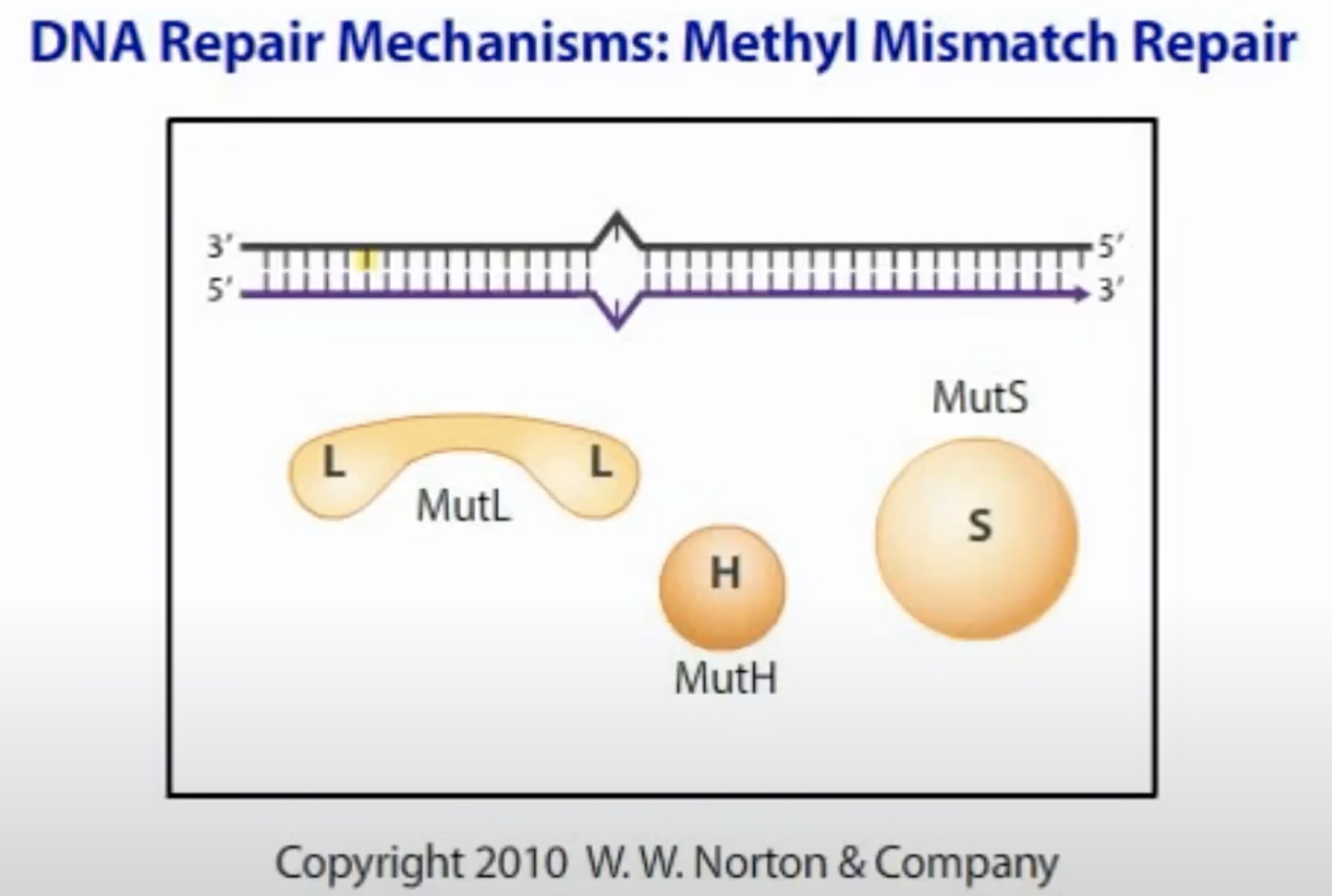
MutS, MutL and MutH add together to repair the mismatch.
-
MutS first bind to the msimatch. Then recruit MutL and MutH
-
MutL recognize the methylated strand and brings it into loop.
-
MutH cleaves the unmethylated strand containing the mutation near the GATC.
-
A DNA helicase called UvrD unwinds the cleave the strand, expose it to a variety of exonuclease. The result is a gap that is filled in by polymerase I.
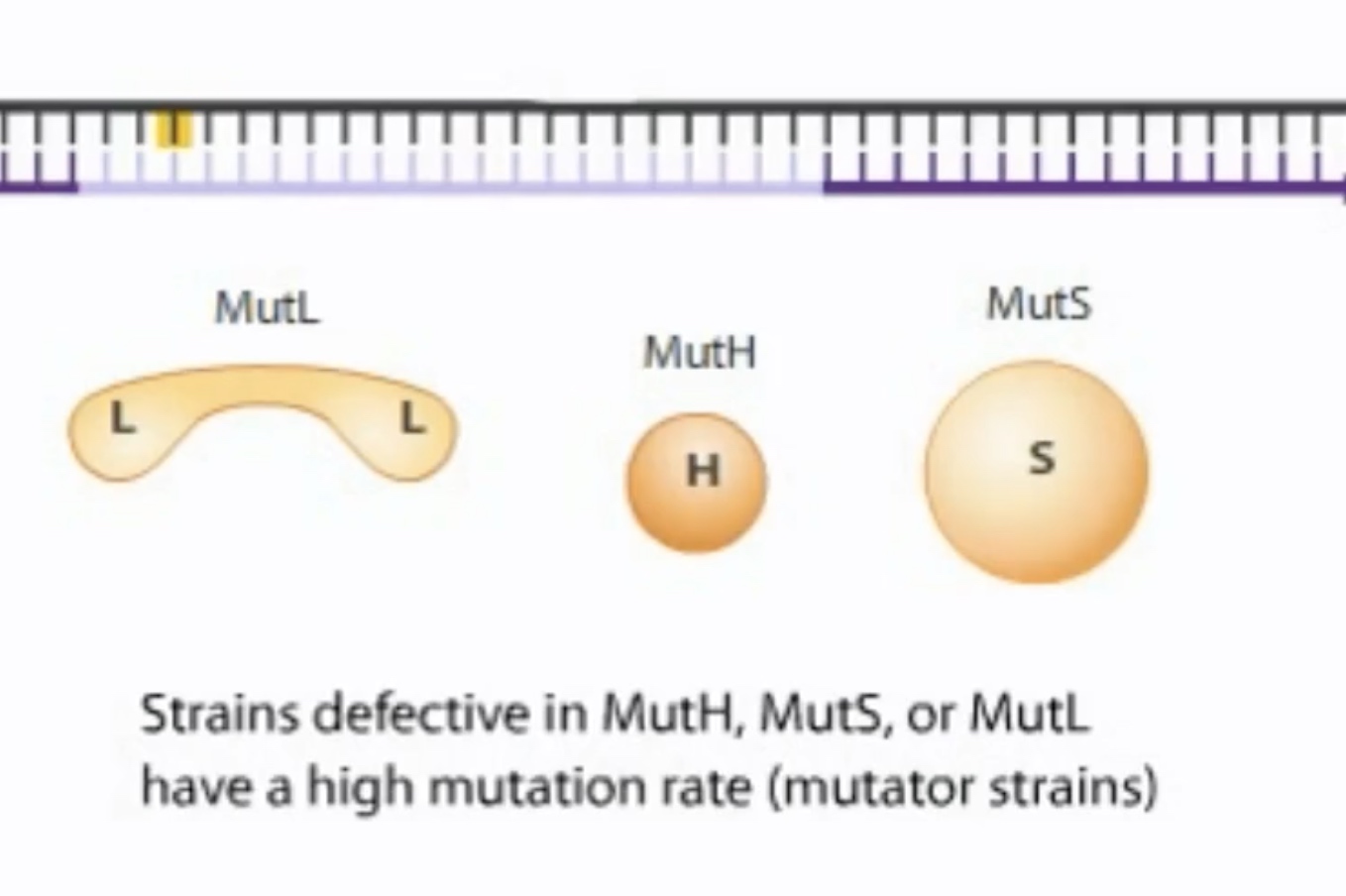
A bacterial strain with a high mutation rate is called a mutator strain.
