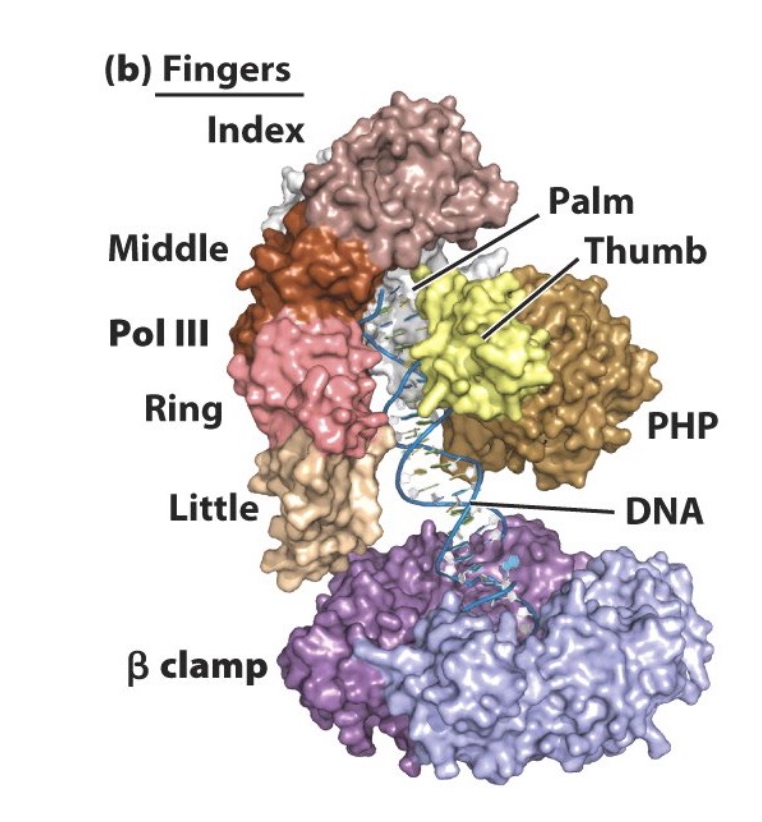
Focus on the proteins that are important to DNA replication process.
The beta clamp position DNA polymerase III at the fork
The DNA double helix is found in the middle of beta clamp.
1. The β clamp increases the processivity of Pol III
The polymerase will dissociate from DNA strand and combine to it again, which takes time and is not efficient.
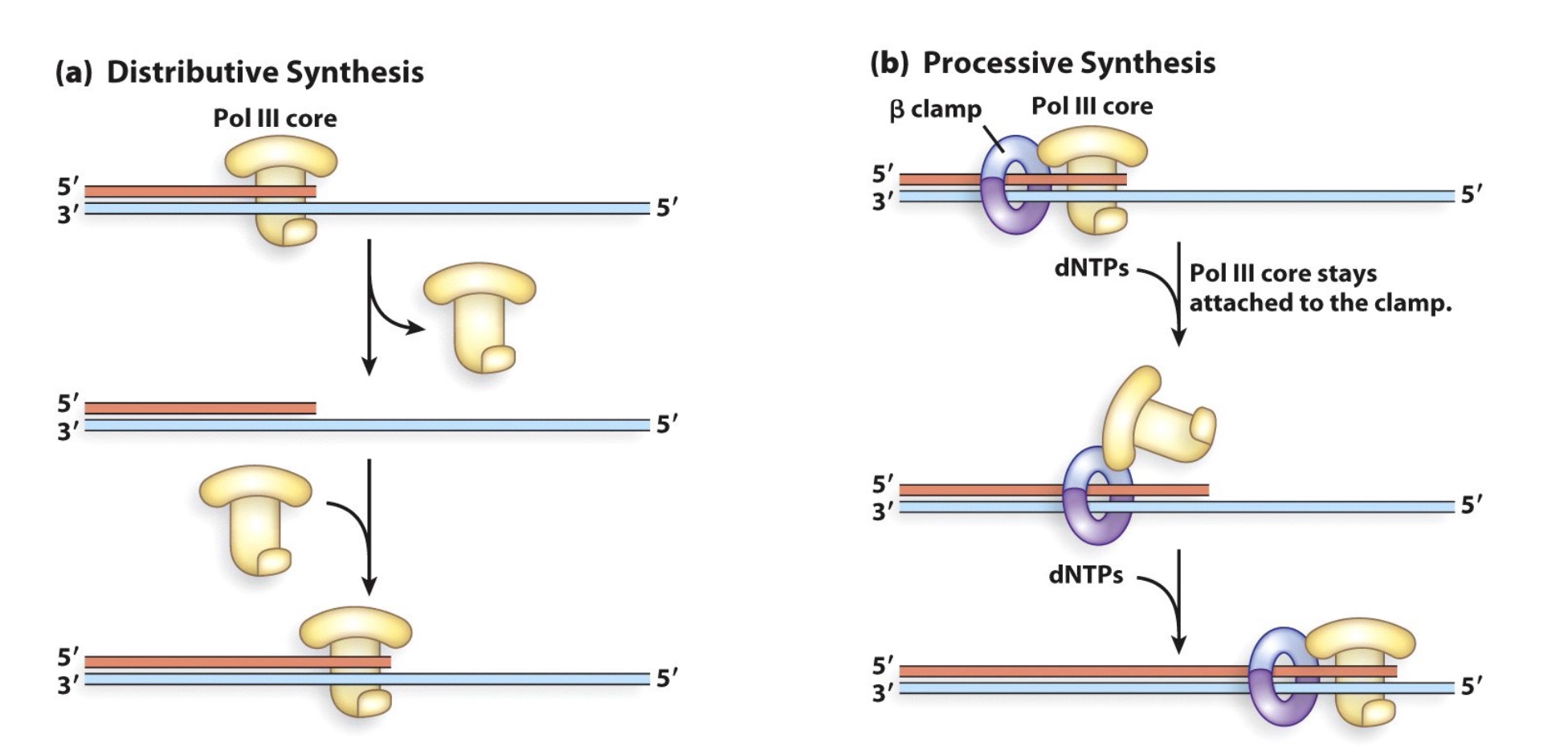
β clamp will help polymerase to combine to the DNA strand. Consequently, makes the replication faster.
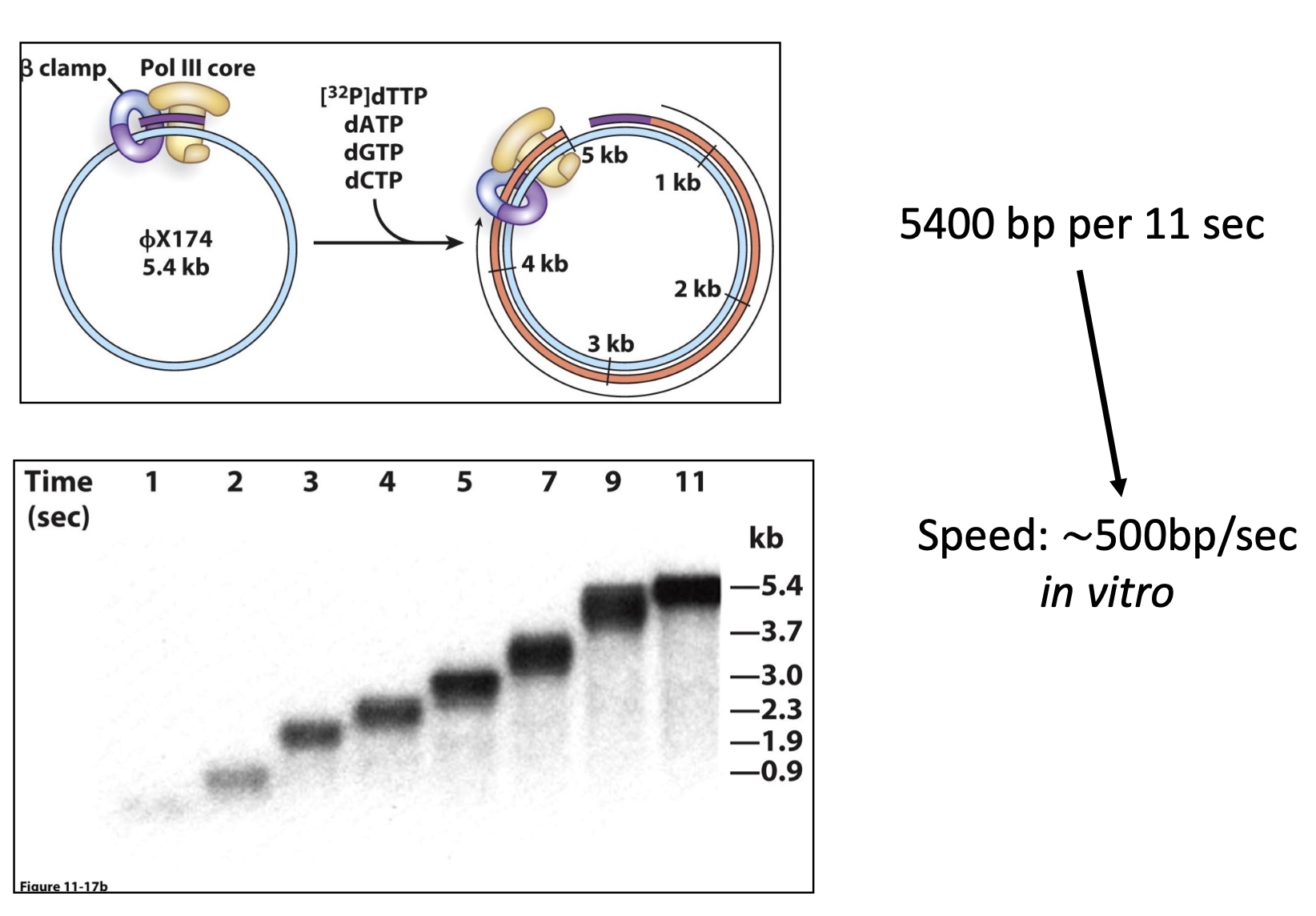
2. The β clamp plays a key role in recruitment of late replication factors(DNA ligase )
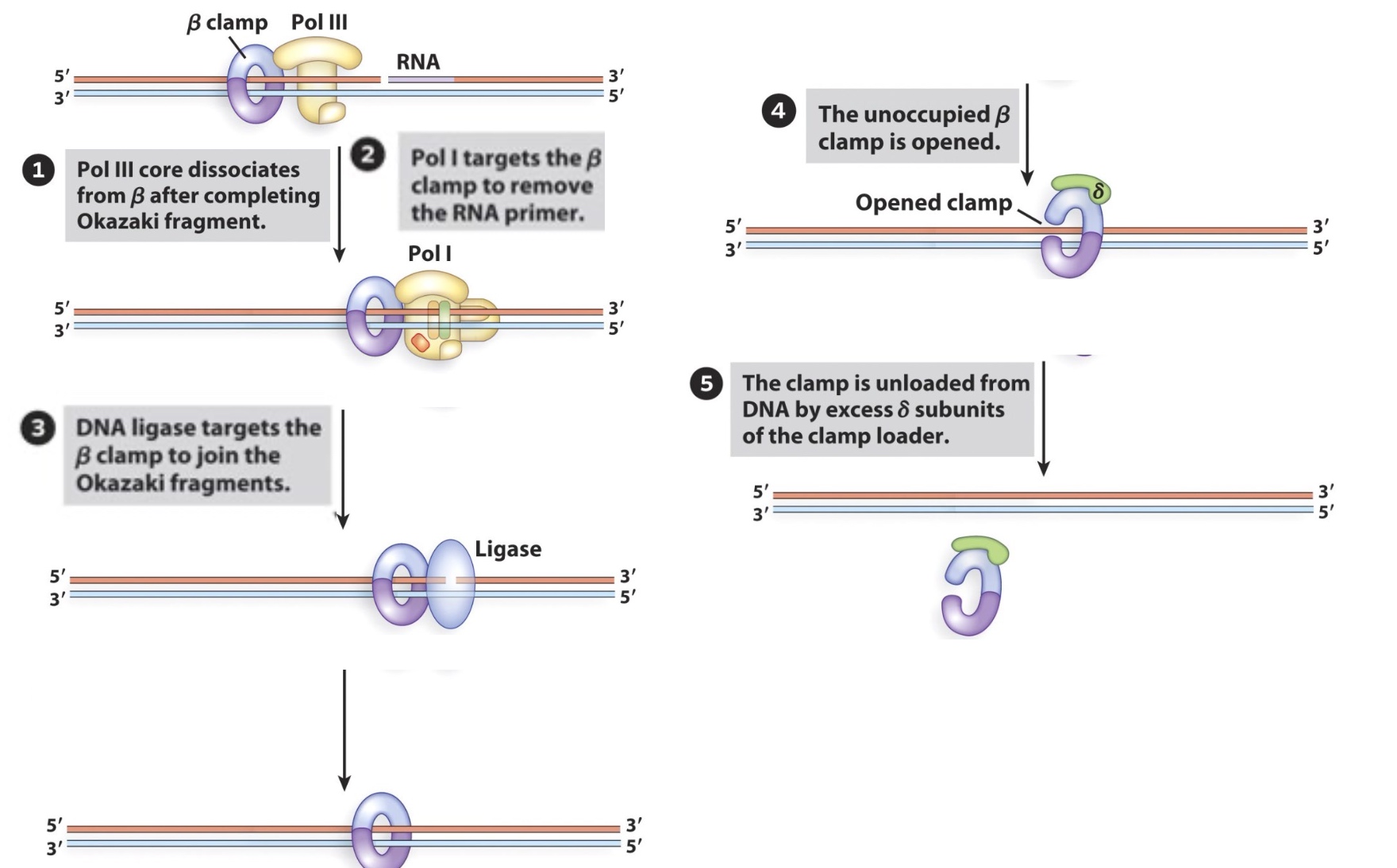
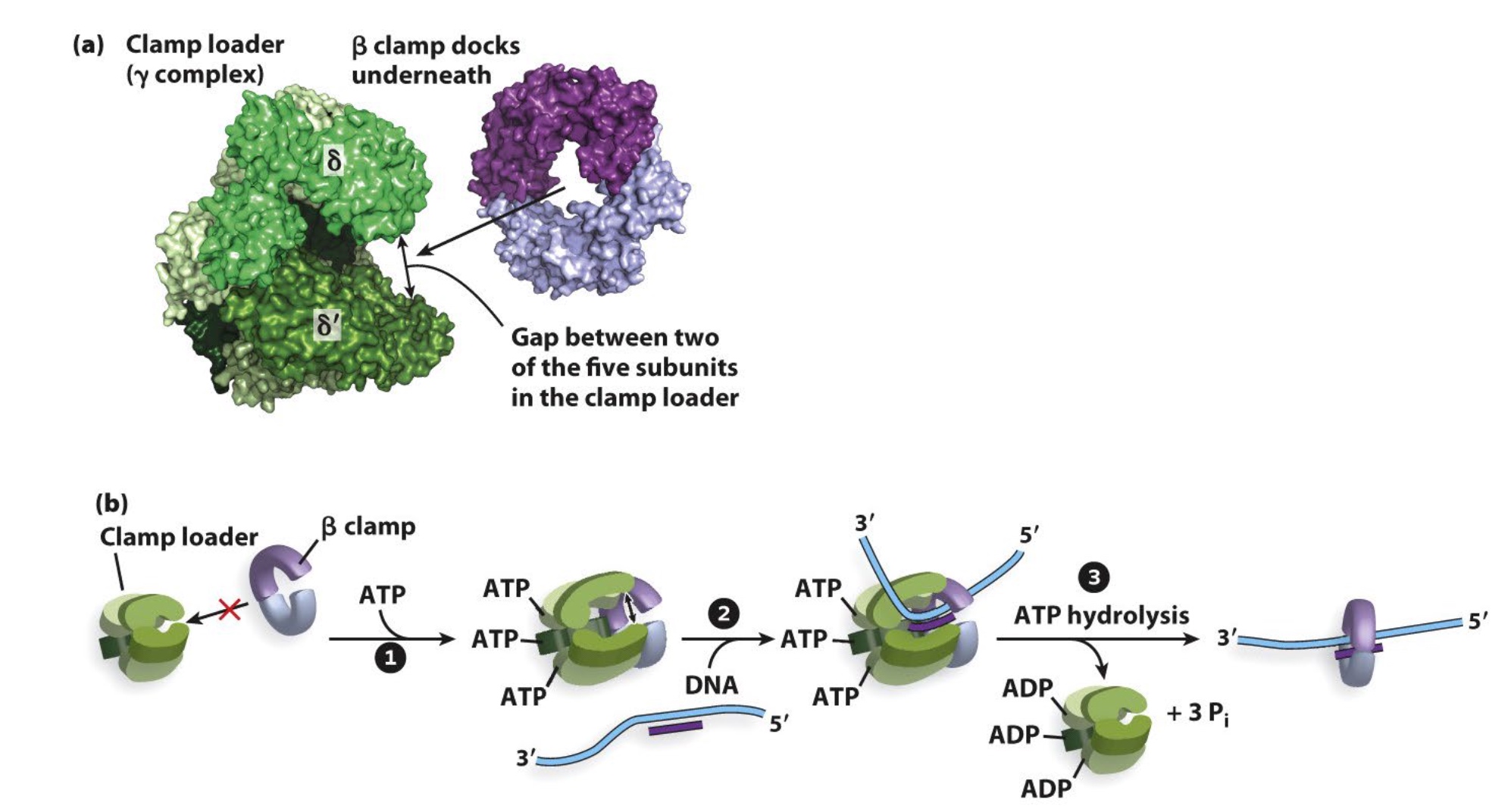
Then clamp loader will join and help open the ring to dissociate clamp from DNA.v
Clamp Loader use ATP to Open and Load Clamps onto dsDNA
dna polymerase does not need ATP.
Like the topoisomerase use the energy stored in triphosphate.
Helicases separate double-stranded nucleic acids and translocate to remove proteins from dsDNA
The unwinding of DNA requires ATP hydrolysis.
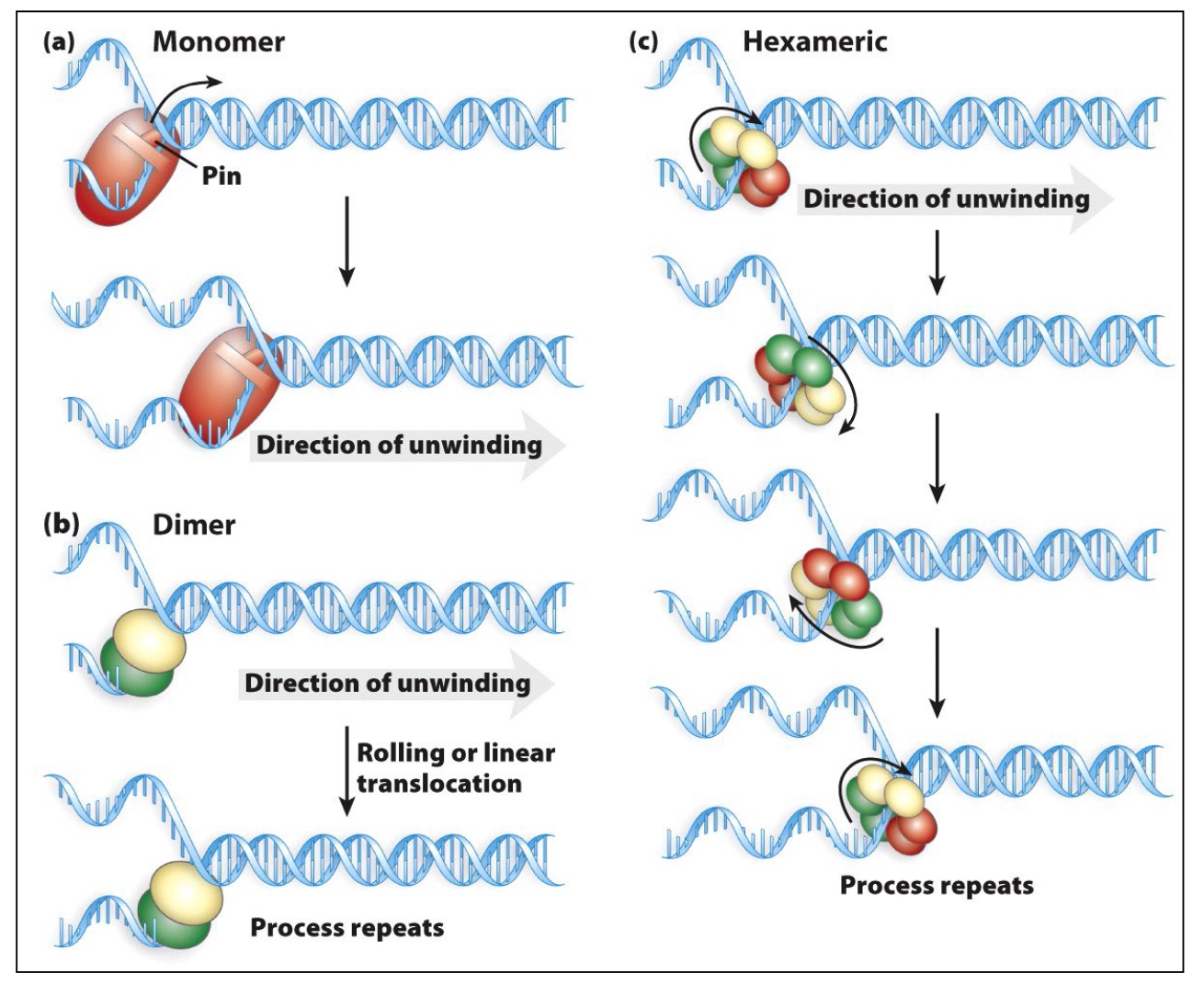
Helicases have directionality. The directionality of helicases is determined based on the movement on the encircled strand.
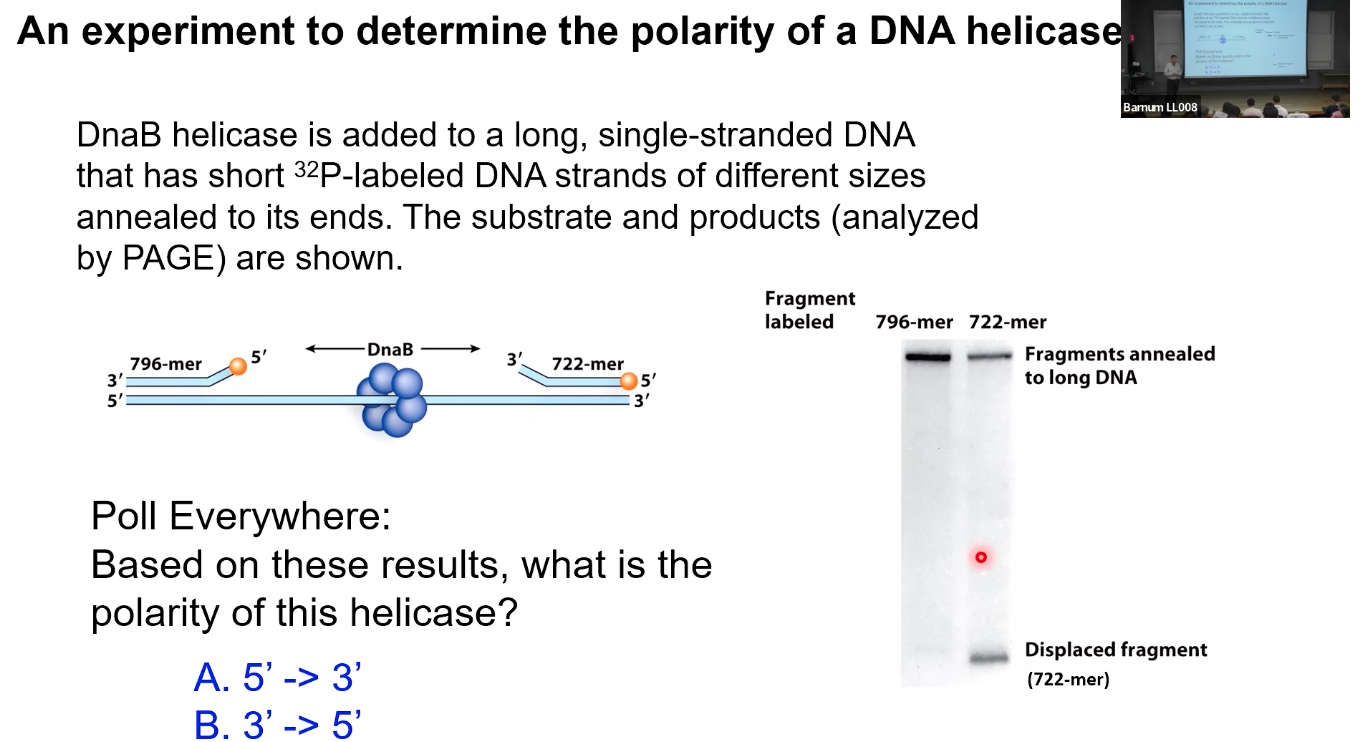
The answer is A.
How does cells control the initiation and termination of DNA replciation?
The bacterial "cell cycle" allows replication to occue continuously
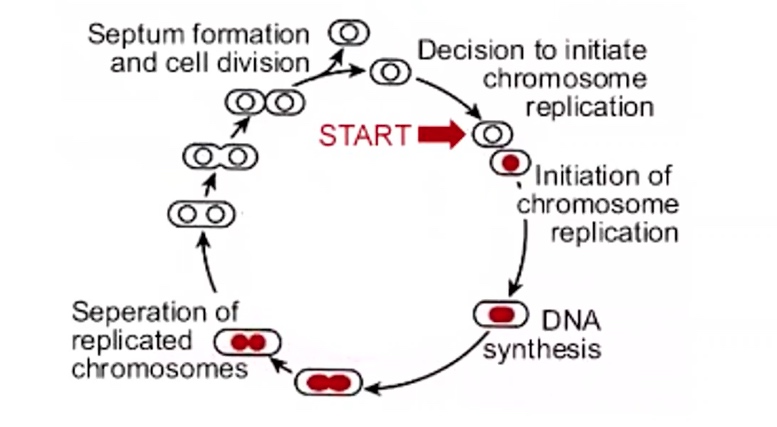
The Eukaryotic cell cycle allows for separation of DNA replication and cell division
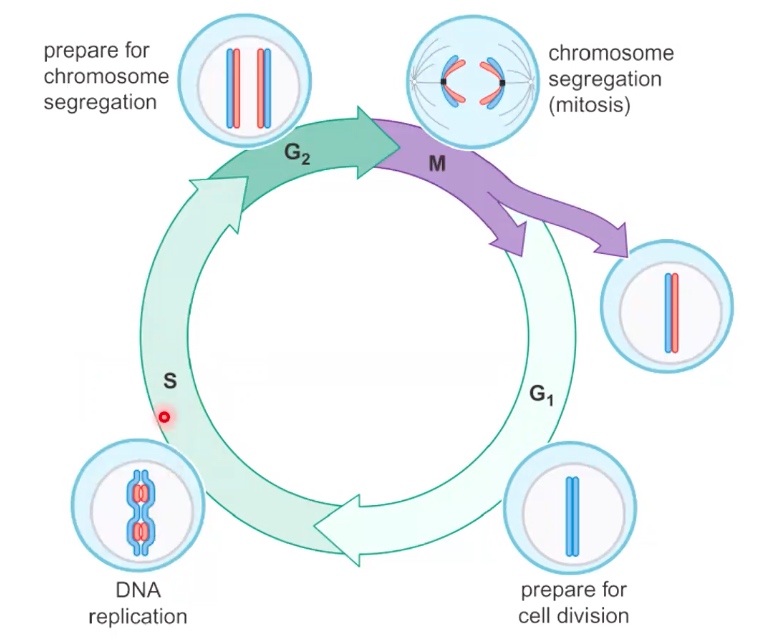
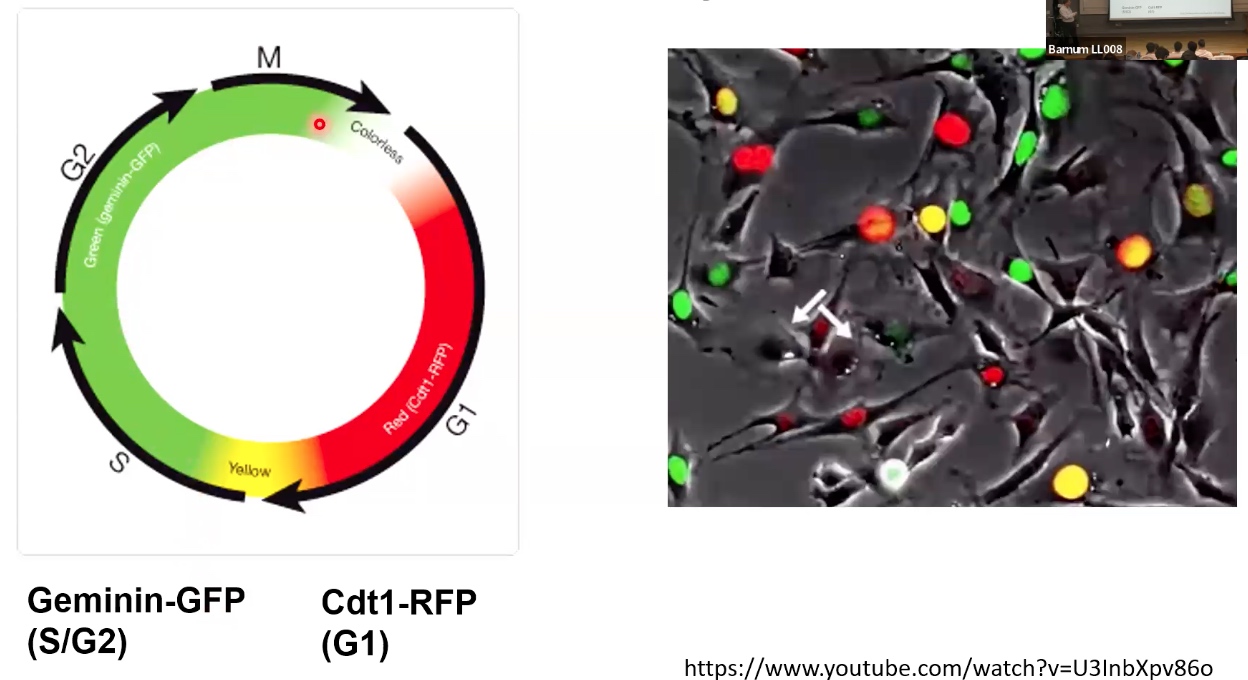
- Start from G1: cells prepare for cell division.
- S: stands for synthesis. It is where DNA replication occurs.
- G2L prepare for cell division and chromosome segregation.
- M: mitosis, where the replicated chromosomes will be separated on the mitotoc spindle.
Strategies for initiation of replication
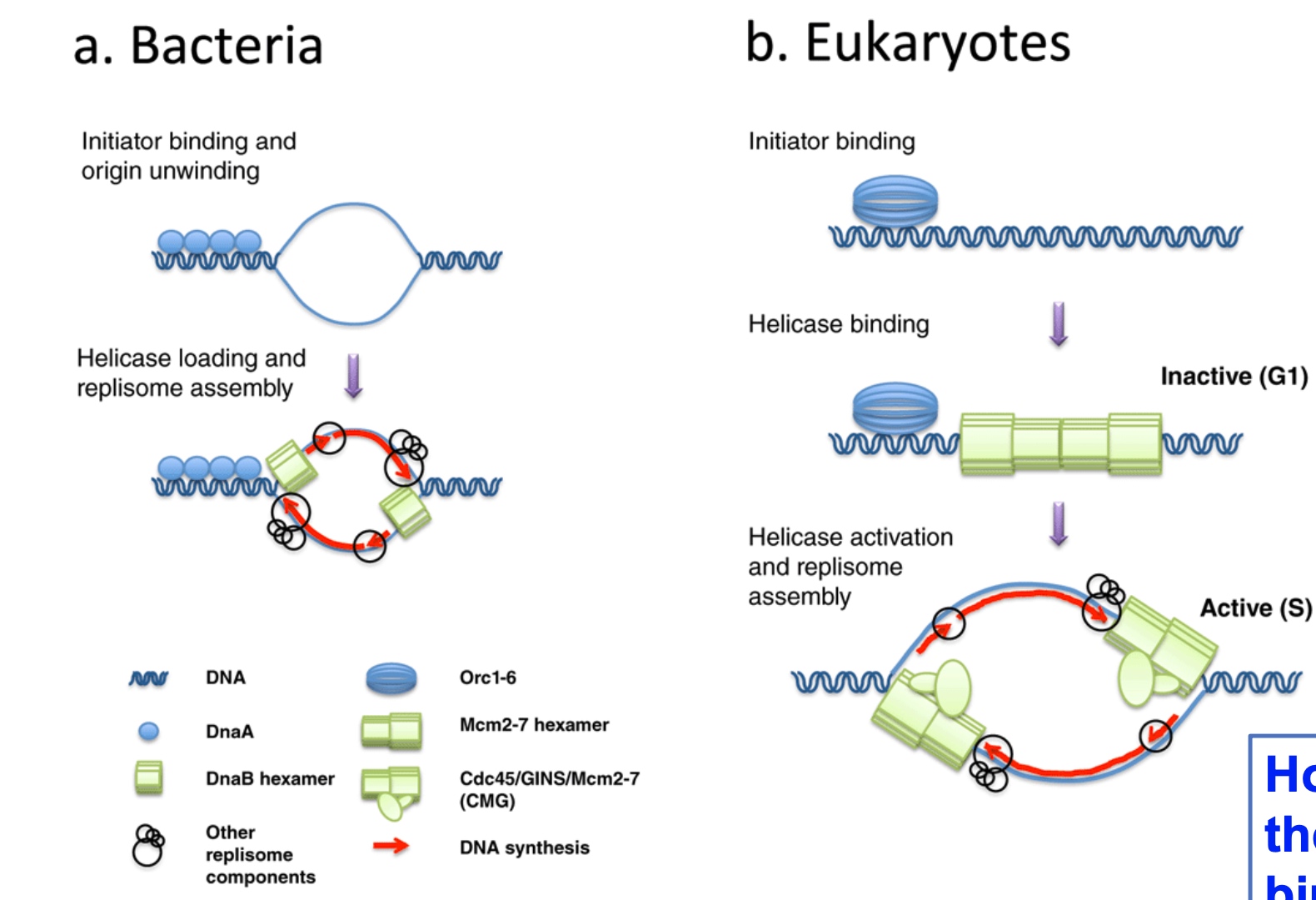
Helicase in eukaryotes does not activate until the signals are received.
How can we identify these initiator bindng sites?
It is important because DNA replication is where cells are the most vulnerable. They are creating a lot of single strabded DNA.
Method1: Genetic Identification of Replicators
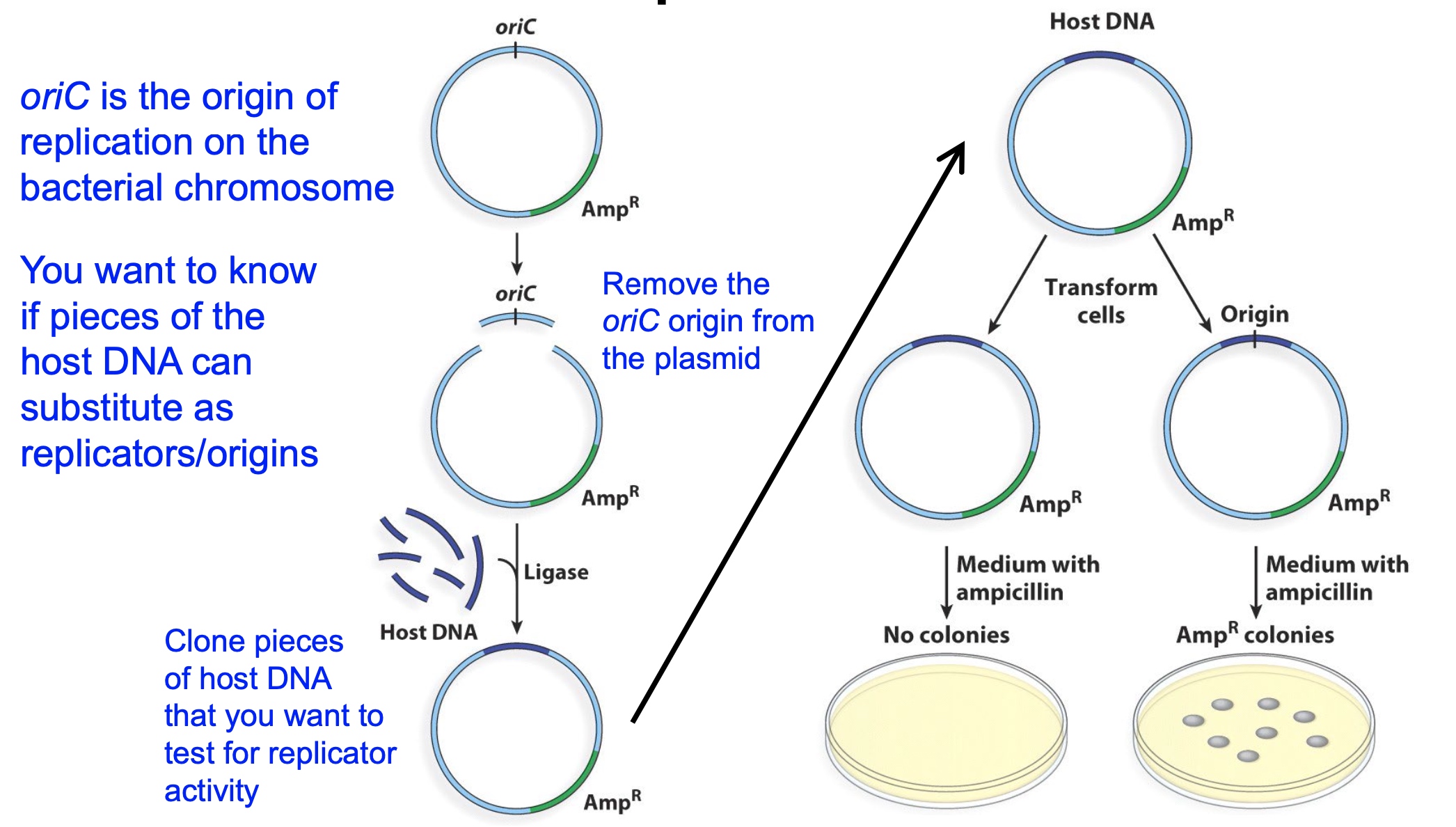
Just substitute the origin of bacteria with host DNA by restriction enzyme to see if host DNAs have the replicator function.
The ampicillin is used to filter out the bacteria with plasmid or not.
858-872 refers to initiator region. Other three lower regions are the points easy to melt.
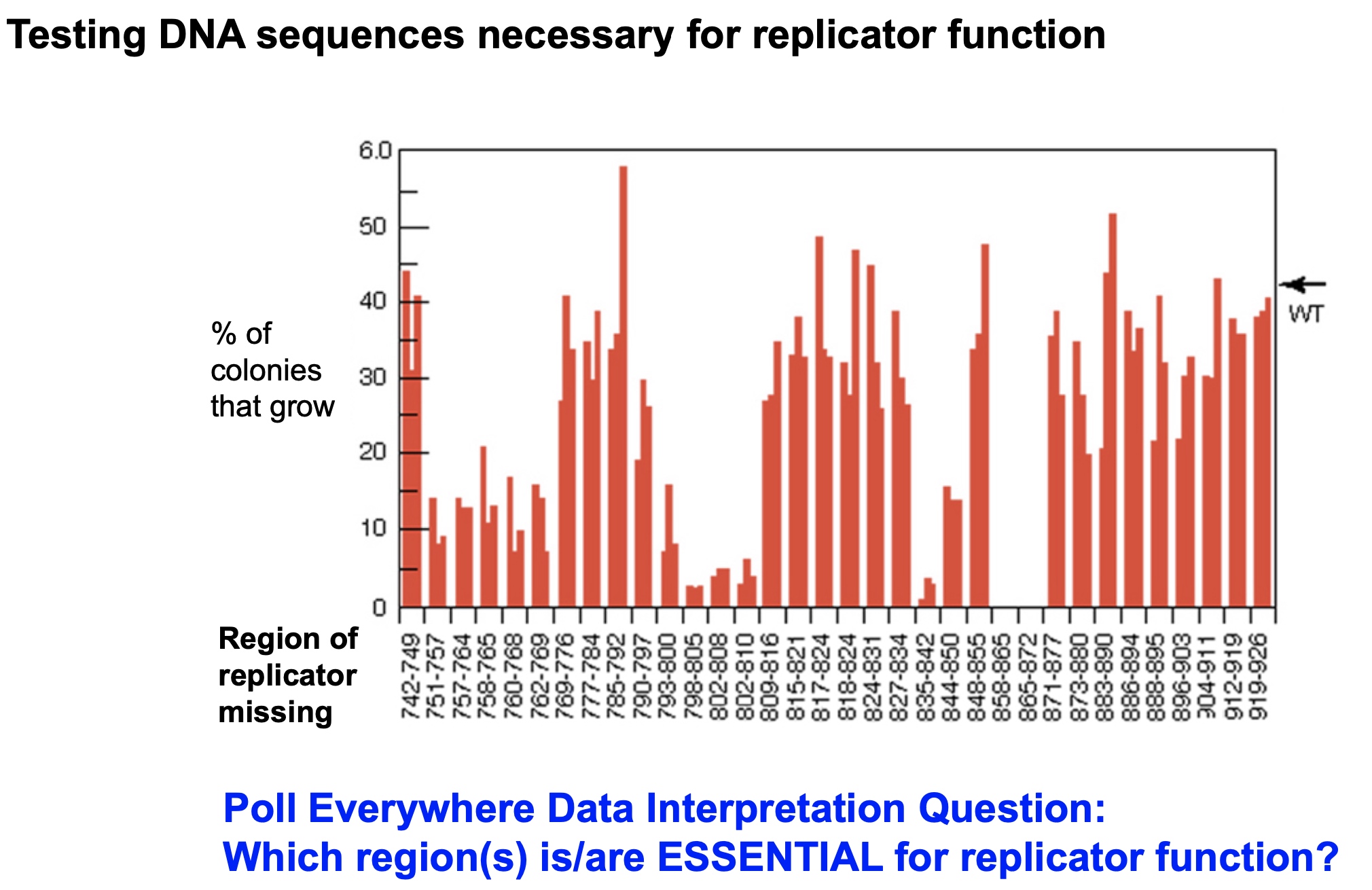
wt means the wild type.
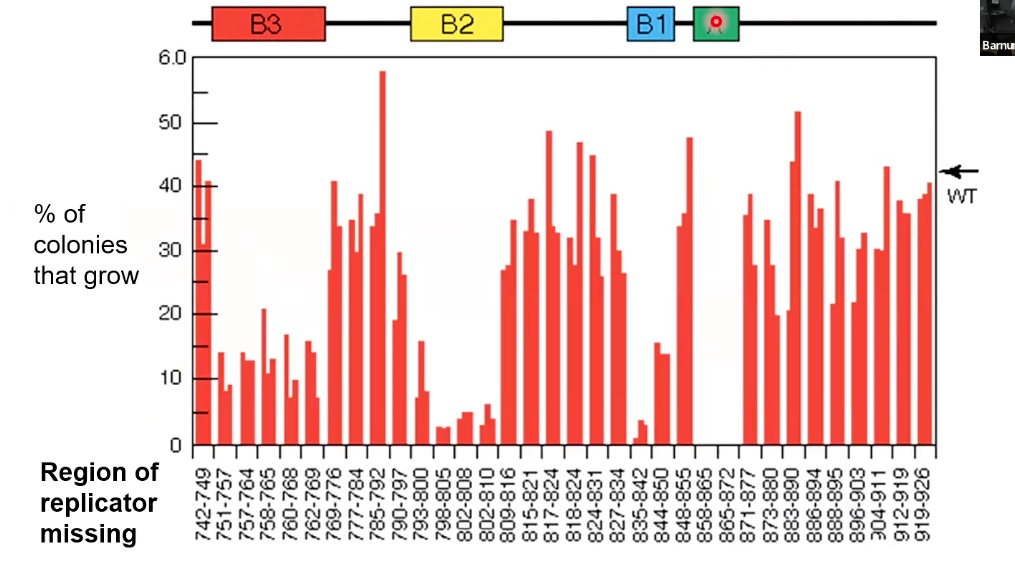
A is the initiator. B1-B3 is important but not essential. B1-B3 may be A-T rich origiin and easy to be melt.
Method2: Physical Detection of Origins: Isolate and Detect Smal Nascent Leading Strands
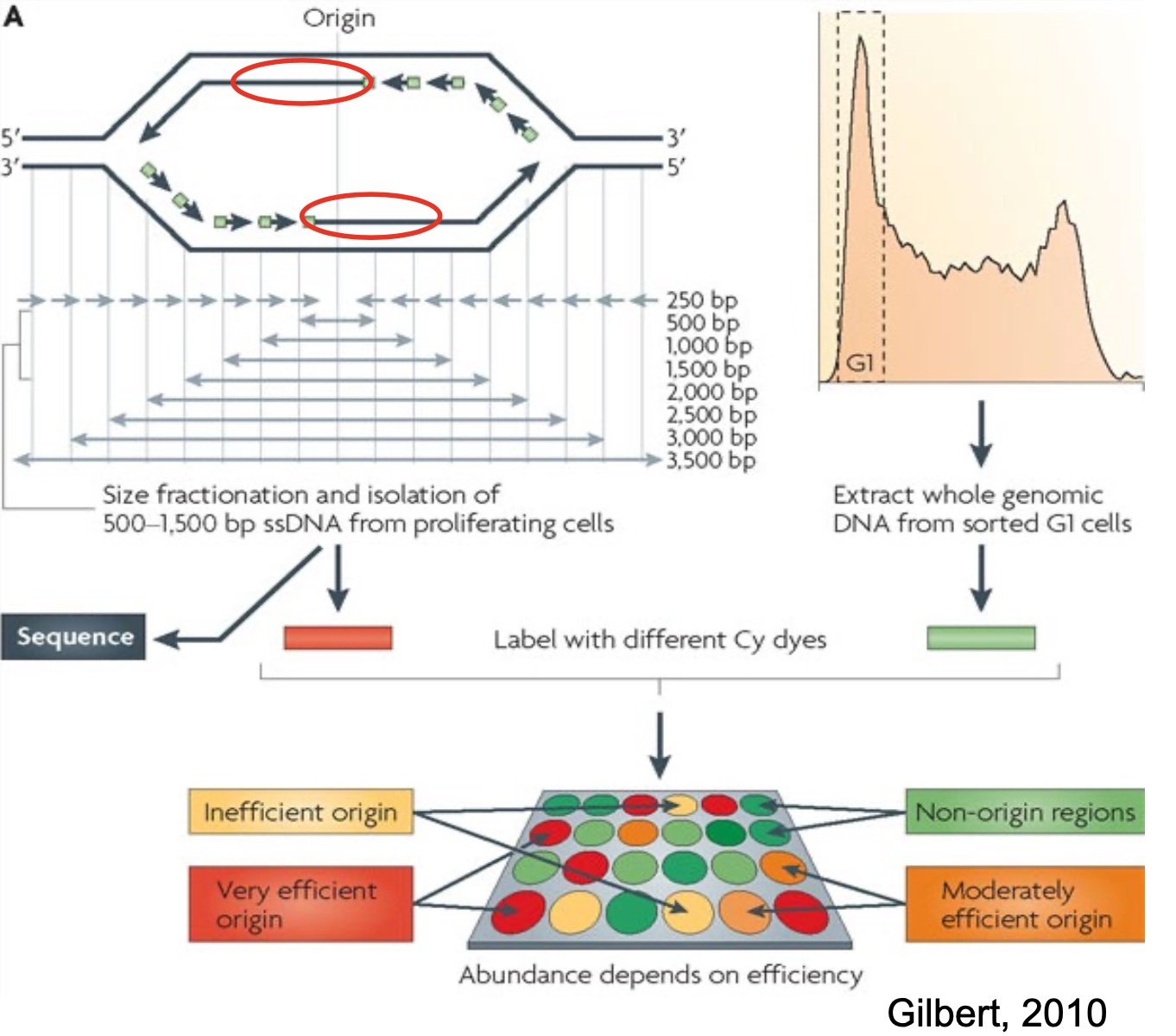
Use NGS or microarray to detect the DNA strand isolated.
Baterial Control of Replication Initiation
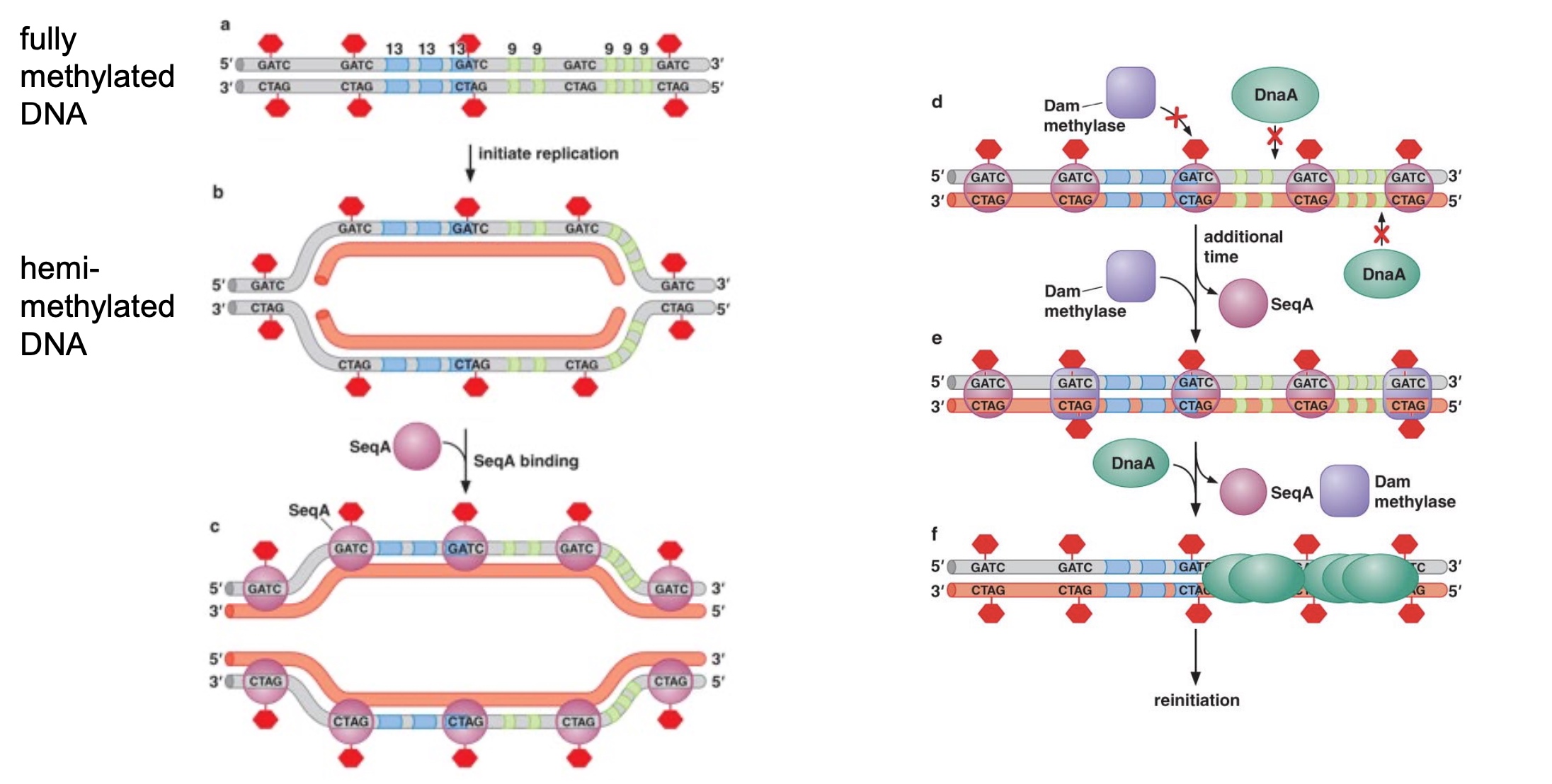
Poll Everywhere synthesis question
If SeqA is mutated so that it has a decreased affinity for hemi-methylated GATC sites, what effect will this have on replication initiation in E. coli?
- A. Replication will initiate faster
- B. Replication will initiate slower
- C. Replication initiation will not be changed
Eukaryotic origins fire only once per cell cycle
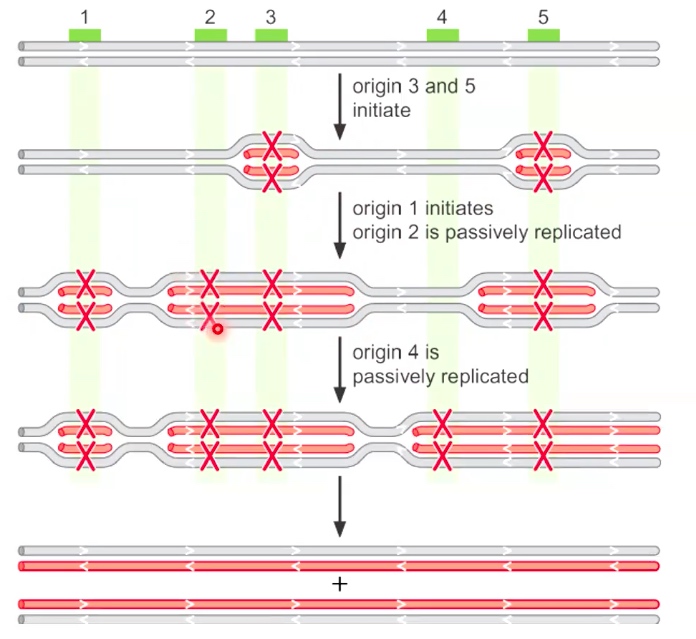
When an origin is marked as red X, which means that origin lost the ability to do replication.
Origin firing: every time initiate the replicatoin.
Initiation of replication in eukaryotes
ORC: Origin recognition complex
Mcm2-7: DNA helicase
Cdt1, Cdc6: Helicase Loader.
CDK, DDK: protein kinase like DDK and CDK will give helicase the signal to do unwinding.
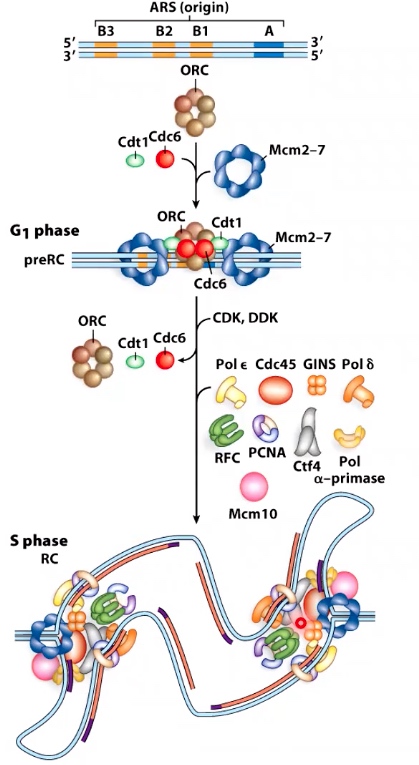
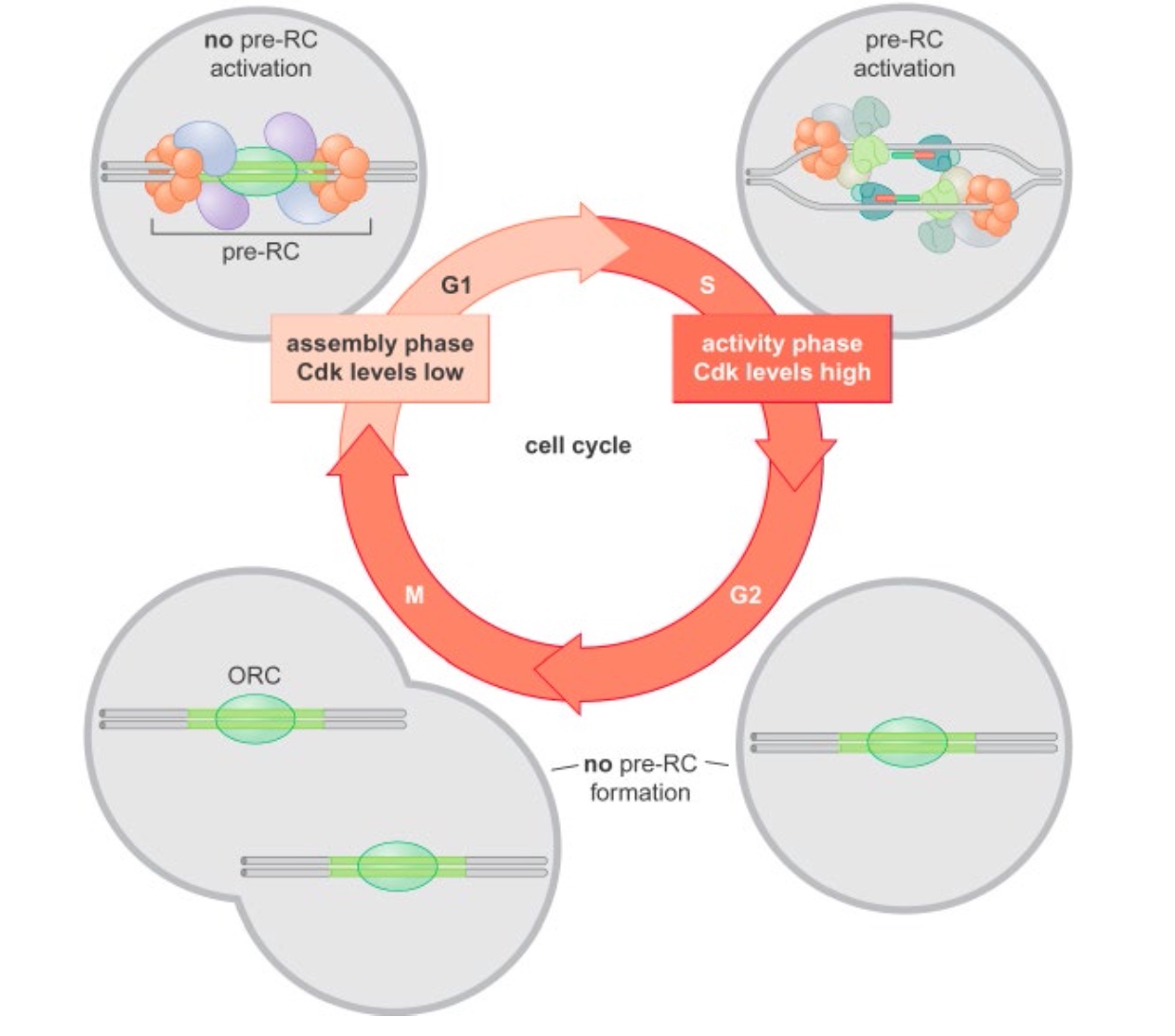
The pre-replicative complex in eukaryotes
The form in G1.
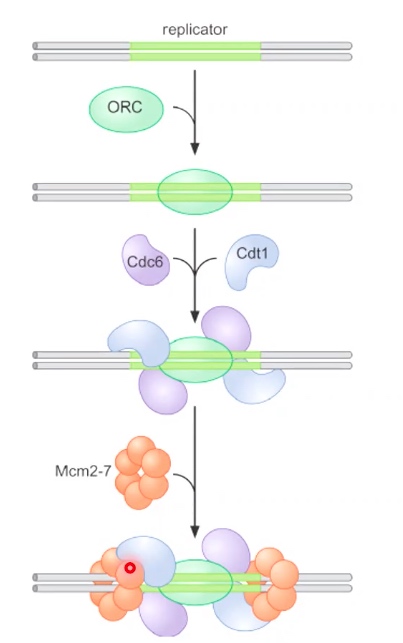
When the cell goes to S phase. DDK, CDK come to give signal.
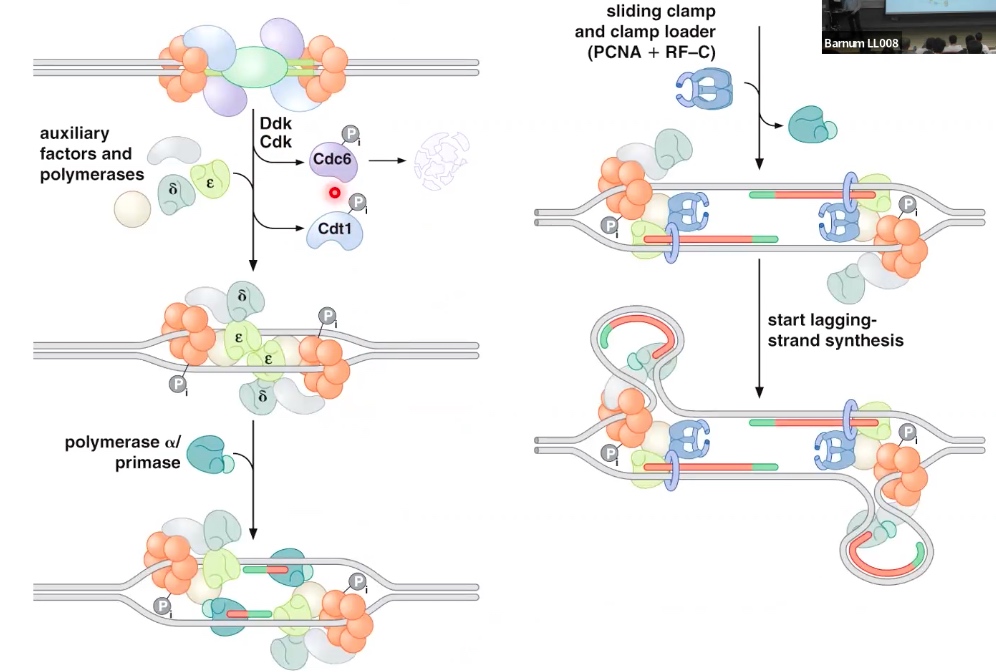
Cyclin dependent kinase(CDK) control the initiation
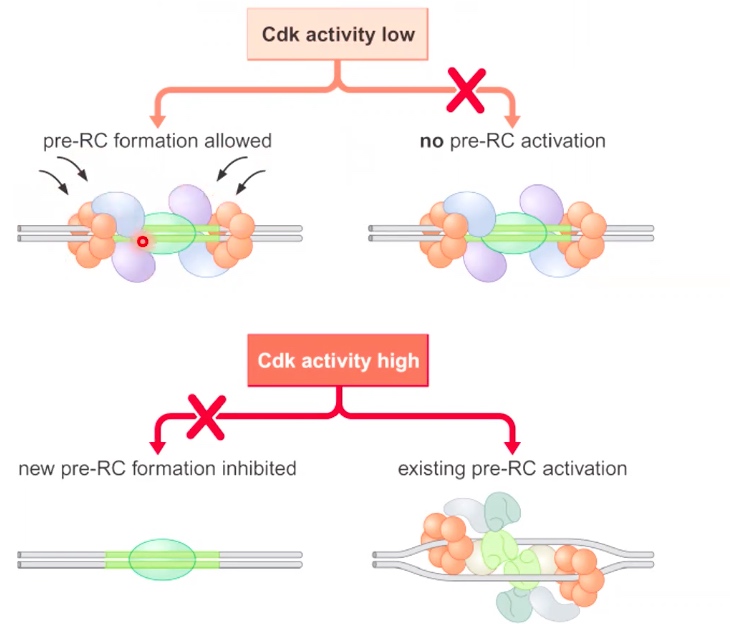
Eukaryotic COntrol of Replication Initiation
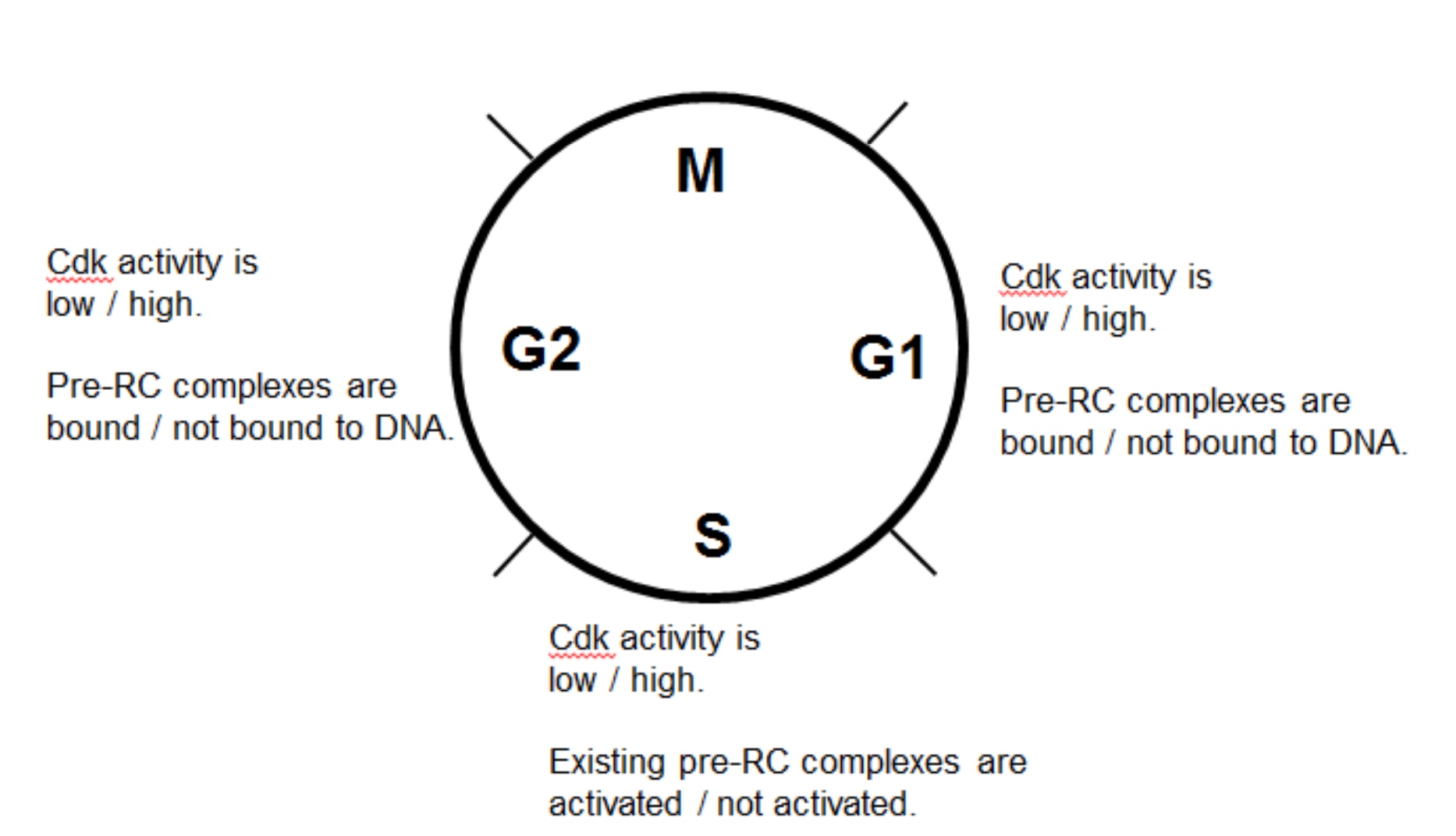
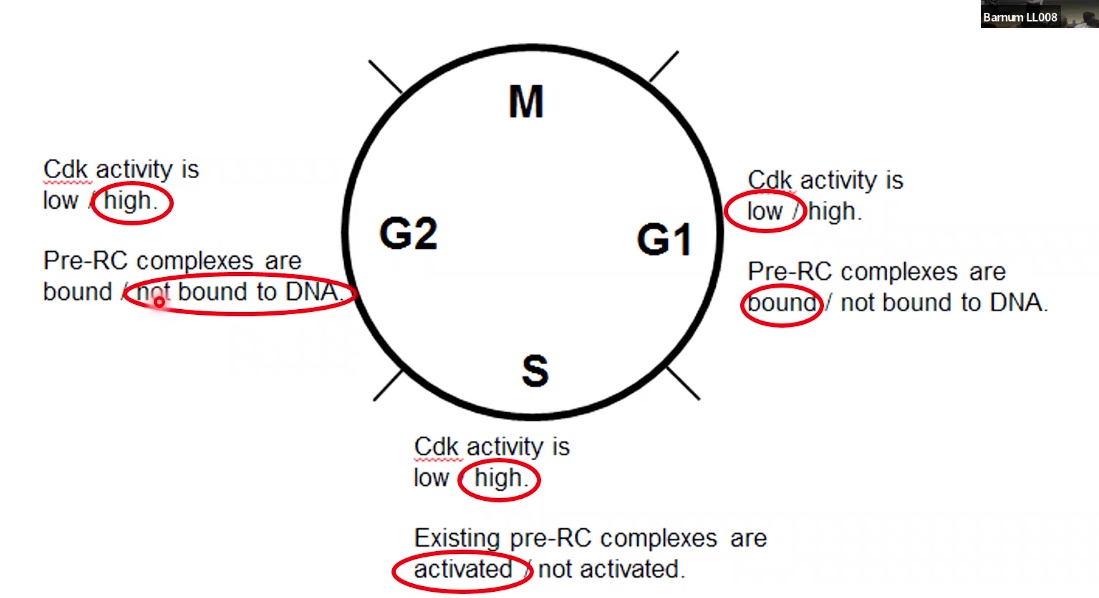
G1: CDK activity is low; PreRC complexes are bound to DNA
S: CDK activity is high; Existing pre-RC complexes are activated
G2: CDK activity is high; PreRC complexes are not bound to DNA.
Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI)
We can fuse these cell cycle regulators to fluorescent proteins.